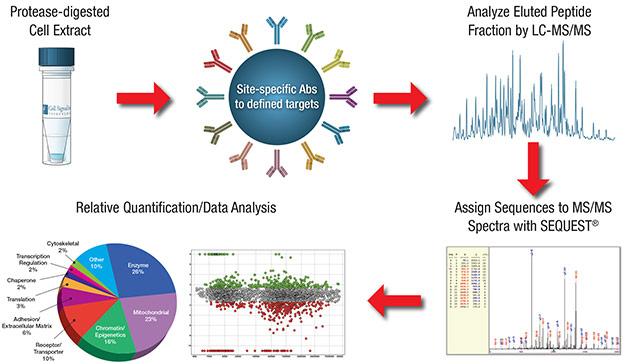
PTMScan® Pathways Services enable identification and quantification of hundreds to thousands of known protein targets within a specific pathway in response to a treatment or disease state. Experienced CST scientists utilize LC/MS based antibody array that combines antibody enrichment of post-translationally modified (PTM) peptides from defined pathways with LC-MS/MS, with complete results obtained in a single run.
You can follow-up on many of the candidate proteins identified via PTMScan® Pathway Services using modification state-specific and total protein antibodies from CST, which are produced and validated in-house for multiple applications.
CST also offers these additional pathway services: