Next generation sequencing (NG-seq) is a high throughput method that can be used downstream of chromatin immunoprecipitation (ChIP) and Cleavage Under Targets and Release Using Nuclease (CUT&RUN) assays to identify and quantify target DNA enrichment across the entire genome. Multiplex Oligos for Illumina (Dual Index Primers) (ChIP-seq, CUT&RUN) contains adaptors and primers that are ideally suited for multiplex sample preparation for NG-seq on the Illumina platform (Illumina, Inc.). This kit can be used to generate up to 96 distinct, barcoded ChIP-seq or CUT&RUN DNA libraries that can be combined into a single sequencing reaction.
Each kit component must pass rigorous quality control standards, and for each new lot the entire set of reagents is functionally validated together by construction and sequencing of indexed libraries on the Illumina sequencing platform.
This product provides enough reagents to support up to 96 DNA sequencing libraries, and must be used in combination with DNA Library Prep Kit for Illumina (ChIP-seq, CUT&RUN) #56795.
Compatible Upstream Assay Kits:
- CUT&RUN Assay Kit #86652
- SimpleChIP® Enzymatic Chromatin IP Kit (Magnetic Beads) #9003
- SimpleChIP® Plus Enzymatic Chromatin IP Kit (Magnetic Beads) #9005
- SimpleChIP® Plus Sonication Chromatin IP Kit (Magnetic Beads) #56383
Non-Compatible Upstream Assay Kits:
Note: Agarose beads are blocked with sonicated salmon sperm DNA, which will contaminate DNA library preps and NG-seq.
- SimpleChIP® Enzymatic Chromatin IP Kit (Agarose Beads) #9002
- SimpleChIP® Plus Enzymatic Chromatin IP Kit (Agarose Beads) #9004
Additional Product Required to Construct DNA Library:
- DNA Library Prep Kit for Illumina (ChIP-seq, CUT&RUN) #56795
Required Reagents:
Reagents Included:
- • (red) Adaptor for Illumina #42436
- • (red) USER Enzyme #59713
- º (white) Index 501 Primer for Illumina #86676
- º (white) Index 502 Primer for Illumina #92596
- º (white) Index 503 Primer for Illumina #29576
- º (white) Index 504 Primer for Illumina #46325
- º (white) Index 505 Primer for Illumina #67343
- º (white) Index 506 Primer for Illumina #89663
- º (white) Index 507 Primer for Illumina #27649
- º (white) Index 508 Primer for Illumina #40566
- • (orange) Index 701 Primer for Illumina #60797
- • (orange) Index 702 Primer for Illumina #79999
- • (orange) Index 703 Primer for Illumina #18697
- • (orange) Index 704 Primer for Illumina #26125
- • (orange) Index 705 Primer for Illumina #39467
- • (orange) Index 706 Primer for Illumina #51808
- • (orange) Index 707 Primer for Illumina #58724
- • (orange) Index 708 Primer for Illumina #65787
- • (orange) Index 709 Primer for Illumina #75272
- • (orange) Index 710 Primer for Illumina #99422
- • (orange) Index 711 Primer for Illumina #10812
- • (orange) Index 712 Primer for Illumina #38569
- Index Primer Orange Tube Caps #54631
- Index Primer White Tube Caps #70942
Reagents Not Included:
- Enzymes and buffers appropriate for ChIP or CUT&RUN Illumina library preparation: provided in DNA Library Prep Kit for Illumina (ChIP-seq, CUT&RUN) #56795
- Nuclease-free Water #12931
- AMPure XP Beads (Beckman Coulter, Inc. #A63881) or SPRIselect Reagent Kit (Beckman Coulter, Inc. #B23317)
- Freshly prepared 80% Ethanol
- 1X TE (10 mM Tris-HCl, pH 8.0, 1 mM EDTA)
- 10 mM Tris-HCl (pH 8.0-8.5)
- Magnetic Separation Rack
- Bioanalyzer and Agilent High Sensitivity DNA Kit (Agilent Technologies, Inc.) PCR tubes and PCR
Multiplex Oligos for Illumina (Dual Index Primers) (ChIP-seq, CUT&RUN) Protocol
I. Low Plexity Pooling Guidelines:
The dual index primer strategy utilizes two 8 base indices within each primer. Index 7 primers contain indices that are adjacent to the P7 sequence while index 5 primers contain indices that are adjacent to the P5 sequence. Dual indexing is enabled by adding a unique index to both ends of a sample to be sequenced. Up to 96 different samples can be uniquely indexed by combining each of the 12 index 7 primers with each of the 8 index 5 primers.
Illumina NG-seq platforms use a red laser/LED to sequence A/C and a green laser/LED to sequence G/T. For each cycle, both the red and the green channel need to be read to ensure proper image registration (i.e. A or C must be present in each cycle, and G or T must be present in each cycle). If this color balance is not maintained, sequencing the index read could fail. Please check the sequences of each index to be used to ensure that you will have signal in both the red and green channels for every cycle. See example below:
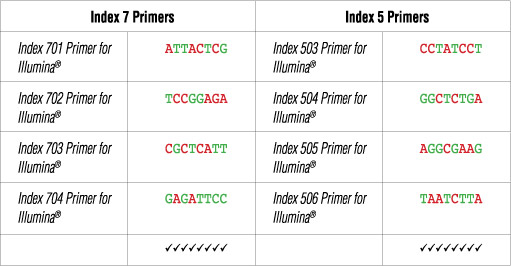
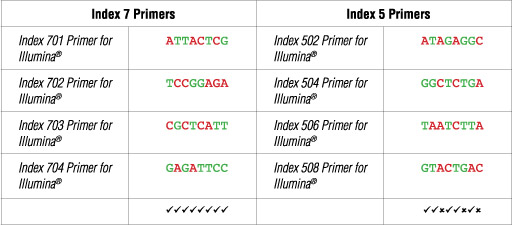
The following table lists some (but not all) valid index combinations that can be sequenced together:

Some other valid combinations are listed below. Choose a valid set of Index 7 primers and a valid set of Index 5 primers. Use each Index 7 primer with each i5 primer to form desired number of primer pairs for PCR amplification of desired number of libraries.

II. Index 5 Primers for Illumina:
Each Index 5 Primer for Illumina is provided in volume of 60 µl.

Where -s- indicates phosphorothioate bond.
III. Index 7 Primers for Illumina:
Each Index 7 Primer for Illumina is provided in a volume of 40 µl.
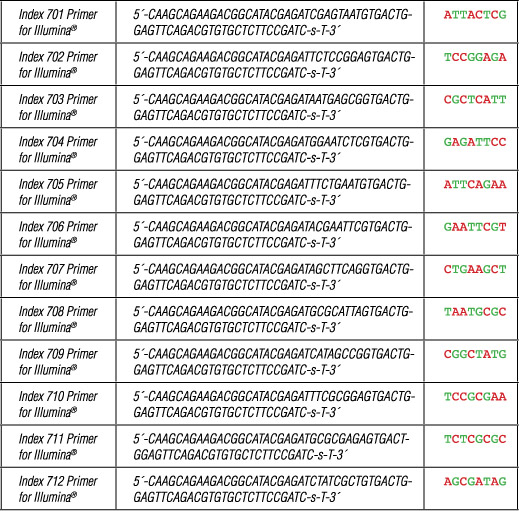
Where -s- indicates phosphorothioate bond.
IV. Set up the PCR Reaction
- Ensure that a valid combination of index 7 and index 5 primers is used. See Section I and II to verify that correct primer combinations have been selected.
- Add only one Index 5 primer (º) (5 µl) and only one Index 7 primer (•) (5 µl) to each PCR tube. It is critical to change tips between tubes to avoid cross-contamination. If necessary, discard the original Index 5 white caps or Index 7 orange caps and apply new caps to avoid index cross-contamination.
- Record the Index 5 and Index 7 primers added to each PCR tube.
- Add 25 µl Q5 PCR Master Mix (•) to each tube that contains primers.
- Add 15 µl of adaptor ligated DNA for a final volume of 50 µl to the corresponding tube. Gently pipette up and down 5–10 times to mix. It is critical to change tips between samples to avoid cross-contamination.
- Record the adaptor ligated DNA sample added to each PCR tube.
- Quickly centrifuge and perform PCR according to recommended cycling conditions (refer to the respective protocols in DNA Library Prep Kit for Illumina (ChIP-seq, CUT&RUN) #56795 for ChIP-DNA or CUT&RUN DNA starting samples).
APPENDIX: Quality Control of the Kit Components
The components in the Multiplex Oligos for Illumina (Dual Index Primers) (ChIP-seq, CUT&RUN) #47538 are individually validated by the functional testing listed below and must pass rigorous quality control standards. Furthermore, each set of components is functionally validated together by construction and sequencing of indexed libraries on the Illumina sequencing platform.
I. Adaptor for Illumina (15 µM) (•)
5´-/5Phos/GAT CGG AAG AGC ACA CGT CTG AAC TCC AGT C/ideoxyU/A CAC TCT TTC CCT ACA CGA CGC TCT TCC GAT C-s-T-3´
Quality Control Assays
- 16-Hour Incubation: 50 µl reactions containing this adaptor and 1 µg of HindIII digested Lambda DNA incubated for 16 hours at 37°C results in no detectable non-specific nuclease degradation as determined by agarose gel electrophoresis. 50 µl reactions containing this reaction buffer at 1X concentration and 1 µg T3 DNA incubated for 16 hours at 37°C also results in no detectable non-specific nuclease degradation as determined by agarose gel electrophoresis.
- Endonuclease Activity: Incubation of a minimum of 5 µl of this adaptor with 1 µg of φX174 RF 1 DNA in assay buffer for 4 hours at 37°C in 50 µl reactions results in < 10% conversion to RF II as determined by agarose gel electrophoresis.
- Phosphatase Activity: Incubation of a minimum of 10 µl of this adaptor in protein phosphatase assay buffer (1 M diethanolamine @ pH 9.8 and 0.5 mM MgCl2) containing 2.5 mM p-nitrophenyl phosphate at 37°C for 4 hours yields no detectable p-nitrophenylene anion as determined by spectrophotometric analysis at 405 nm.
- RNase Activity: Incubation of this adaptor with 40 ng of a FAM-labeled RNA transcript for 16 hours at 37°C results in no detectable RNase activity as determined by polyacrylamide gel electrophoresis.
II. USER Enyzme (•)
Supplied in: 50 mM KCl, 5 mM NaCl, 10 mM Tris-HCl (pH 7.4 @ 25°C), 0.1 mM EDTA, 1 mM DTT, 175 µg/ml BSA, and 50% Glycerol
Quality Control Assays
- Non-Specific DNase Activity (16 Hour): A 50 µl reaction in NEBuffer 1 containing 1 µg of Lambda DNA and a minimum of 50 units of Uracil DNA Glycosylase incubated for 16 hours at 37°C results in a DNA pattern free of detectable nuclease degradation as determined by agarose gel electrophoresis. A 50 µl reaction in Endonuclease VIII Reaction Buffer containing 1 µg of Lambda-HindIII DNA and a minimum of 25 units of Endonuclease VIII incubated for 16 hours at 37°C results in a DNA pattern free of detectable nuclease degradation as determined by agarose gel electrophoresis.
- Exonuclease Activity (Radioactivity Release): A 50 µl reaction in NEBuffer 1 containing 1 µg of a mixture of single and double-stranded [3H] E. coli DNA and a minimum of 50 units of Uracil DNA Glycosylase incubated for 4 hours at 37°C releases < 0.1% of the total radioactivity. A 50 µl reaction in Endonuclease VIII Reaction Buffer containing 1 µg of a mixture of single and double-stranded [3H] E. coli DNA and a minimum of 10 units of Endonuclease VIII incubated for 4 hours at 37°C releases < 0.5% of the total radioactivity.
- Endonuclease Activity (Nicking): A 50 µl reaction in UDG Reaction Buffer containing 1 µg of supercoiled φX174 DNA and a minimum of 50 units of Uracil DNA Glycosylase incubated for 4 hours at 37°C results in < 10% conversion to the nicked form as determined by agarose gel electrophoresis.
- Phosphatase Activity: Incubation of a minimum of 10 µl of USER at a 1X concentration in protein phosphatase assay buffer (1 M diethanolamine @ pH 9.8 and 0.5 mM MgCl2) containing 2.5 mM p-nitrophenyl phosphate at 37°C for 4 hours yields no detectable p-nitrophenylene anion as determined by spectrophotometric analysis at 405 nm.
III. Index 5 and Index 7 Primers for Illumina (10 µM) (º•)
Quality Control Assays
- 16-Hour Incubation: 50 µl reactions containing 1 µl Index [X] Primer for Illumina and 1 µg of HindIII digested Lambda DNA incubated for 16 hours at 37°C results in no detectable non-specific nuclease degradation as determined by agarose gel electrophoresis. 50 µl reactions containing Index [X] Primer for Illumina and 1 µg of T3 DNA incubated for 16 hours at 37°C results in no detectable non-specific nuclease degradation as determined by agarose gel electrophoresis.
- Endonuclease Activity: Incubation of a 50 µl reaction containing 1 µl Index [X] Primer for Illumina with 1 µg of φX174 RF I supercoiled DNA for 4 hours at 37°C results in less than 10% conversion to RF II (nicked molecules) as determined by agarose gel electrophoresis.
- RNase Activity: Incubation of a 10 µl reaction containing 1 µl Index [X] Primer for Illumina with 40 ng of RNA transcript for 16 hours at 37°C resulted in no detectable degradation of RNA as determined by gel electrophoresis.
- Phosphatase Activity: Incubation of Index [X] Primer for Illumina in protein phosphatase assay buffer (1 M diethanolamine @ pH 9.8 and 0.5 mM MgCl2) containing 2.5 mM p-nitrophenyl phosphate at 37°C for 4 hours yields no detectable p-nitrophenylene anion as determined by spectrophotometric analysis at 405 nm.





