CST signaling pathway diagrams allow you to click on individual nodes to find research resources or product information. You can also download the pathway diagrams for educational and research purposes.
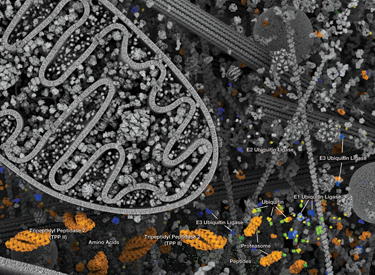
The ubiquitin-proteasome system (UPS) is the primary means by which cellular proteins are degraded and is a highly regulated system for elimination of misfolded or damaged proteins as well as proteins whose activity is acutely regulated by signaling pathways. This system plays a central role in cell proliferation, transcriptional regulation, apoptosis, immunity, and development. Ubiquitin is a highly conserved 76-amino acid protein that can be covalently linked to many cellular proteins by the ubiquitination process. Ubiquitination is an ATP-dependent process carried out by three classes of enzymes. A "ubiquitin activating enzyme" (E1) forms a thio-ester bond with ubiquitin. This reaction allows subsequent binding of ubiquitin to a "ubiquitin conjugating enzyme" (E2), followed by the formation of an isopeptide bond between the carboxy-terminus of ubiquitin and the ε-amino group of a lysine residue on the substrate protein. The latter reaction requires a "ubiquitin ligase" (E3). Several hundred E3 ligases exist within the eukaryotic cell; each ligase can only modify a subset of substrate proteins, thereby providing substrate specificity to the system. Ubiquitinated proteins are then targeted to the 26S proteasome for degradation or experience changes in protein location or activity.
Ubiquitin can be linked to a substrate as a single unit, monoubiquitination, or as a branched chain, polyubquitination. Substrate proteins are linked to ubiquitin using seven distinct ubiquitin lysine residues (Lys6, Lys11, Lys27, Lys29, Lys33, Lys48 and Lys63). Formation of a polyubiquitin chain occurs when a lysine residue of ubiquitin is linked to the carboxy-terminal glycine of another ubiquitin. Polyubiquitinated proteins have distinct fates depending upon the nature of the ubiquitin linkage through which they are conjugated; K48-linked polyubiquitin chains mainly target proteins for proteasomal degradation while K63-linked polyubiquitin typically regulates protein function, subcellular localization, and protein-protein interactions, although this linkage sometimes results in proteasomal degradation as well.
Deubiquitinating enzymes (DUBs) reverse the process of ubiquitination by removing ubiquitin from its substrate protein. DUB activity maintains ubiquitin recycling and ensures the cellular pool of ubiquitin molecules remains steady. DUBs are categorized into 5 subfamilies: USP, UCH, OTU, MJD, and JAMM, each with a specific tissue and substrate specificity. The 26S proteasome is a highly abundant ~2 MDa complex that serves as the proteolytic arm of the ubiquitin-proteasome system. It consists largely of two sub-complexes, the 19S regulatory particle (RP) and the 20S catalytic core particle (CP). The CP forms a cylindrical catalytic chamber. The RP includes a base and a lid. The base functions to unfold the substrate and open the gate, thus allowing entry of the unfolded substrate into the catalytic chamber. The lid is predominantly involved in specific recognition of the ubiquitin signal. In addition to the 19S cap, other proteins and complexes bind to the end of the 20S cylinder and activate it by facilitating opening of the gate. Furthermore, proteasome associated DUBs and E3s can remodel substrate-anchored polyubiquitin chains, which may modulate their susceptibility to degradation.
Small ubiquitin-related modifier 1, 2, and 3 (SUMO-1, -2, and -3) and NEDD8 are members of the ubiquitin-like protein family. SUMO and NEDD can be covalently attached to proteins (termed sumoylation and neddylation, respectively) in a manner analogous to ubiquitination using an E1, E2, E3 conjugation system. Unlike ubiquitination, however, sumoylation and neddylation of substrate proteins do not typically result in degradation. Instead, the SUMO and NEDD modifications affect subcellular localization, protein function, or protein-protein interactions. Sumoylation has numerous cellular effects including nuclear trafficking, regulation of transcriptional activity, and protein stability. The cross-talk between sumoylation and other post-translational modifications such as ubiquitination, phosphorylation, and acetylation is an area of active research.