| Cat. # | Size | Qty. | Price |
|---|---|---|---|
| 29580S | 1 Kit (24 assays) |
|
| Product Includes | Volume (with Count) | Storage Temp | |||
|---|---|---|---|---|---|
| Adaptor for Illumina Systems | 1 x 240 µl | -20°C | |||
| USER Enzyme | 1 x 72 µl | -20°C | |||
| Universal PCR Primer for Illumina Systems | 1 x 120 µl | -20°C | |||
| Index 1 Primer for Illumina Systems | 1 x 10 µl | -20°C | |||
| Index 2 Primer for Illumina Systems | 1 x 10 µl | -20°C | |||
| Index 3 Primer for Illumina Systems | 1 x 10 µl | -20°C | |||
| Index 4 Primer for Illumina Systems | 1 x 10 µl | -20°C | |||
| Index 5 Primer for Illumina Systems | 1 x 10 µl | -20°C | |||
| Index 6 Primer for Illumina Systems | 1 x 10 µl | -20°C | |||
| Index 7 Primer for Illumina Systems | 1 x 10 µl | -20°C | |||
| Index 8 Primer for Illumina Systems | 1 x 10 µl | -20°C | |||
| Index 9 Primer for Illumina Systems | 1 x 10 µl | -20°C | |||
| Index 10 Primer for Illumina Systems | 1 x 10 µl | -20°C | |||
| Index 11 Primer for Illumina Systems | 1 x 10 µl | -20°C | |||
| Index 12 Primer for Illumina Systems | 1 x 10 µl | -20°C |
Product Information
Next generation sequencing (NG-seq) is a high throughput method that can be used downstream of chromatin immunoprecipitation (ChIP) and Cleavage Under Targets and Release Using Nuclease (CUT&RUN) assays to identify and quantify target DNA enrichment across the entire genome. Multiplex Oligos for Illumina Systems (Single Index Primers) (ChIP-seq, CUT&RUN) contain adaptors and primers that are ideally suited for multiplex sample preparation for NG-seq on the Illumina Systems platform (Illumina, Inc.). This kit can be used to generate up to 12 distinct, barcoded ChIP-seq or CUT&RUN DNA libraries that can be combined into a single sequencing reaction.
Each kit component must pass rigorous quality control standards, and for each new lot the entire set of reagents is functionally validated together by construction and sequencing of indexed libraries on the Illumina Systems sequencing platform.
This product provides enough reagents to support up to 24 DNA sequencing libraries, and must be used in combination with the DNA Library Prep Kit for Illumina Systems (ChIP-seq, CUT&RUN) #56795.
Compatible Assay kits:
SimpleChIP® Enzymatic Chromatin IP Kit (Magnetic Beads) #9003
SimpleChIP® Plus Enzymatic Chromatin IP Kit (Magnetic Beads) #9005
SimpleChIP® Plus Sonication Chromatin IP Kit #56383
DNA Library Prep Kit for Illumina Systems (ChIP-seq, CUT&RUN) #56795
CUT&RUN Assay Kit #86652
Non-Compatible SimpleChIP® kits:
SimpleChIP® Enzymatic Chromatin IP Kit (Agarose Beads) #9002
SimpleChIP® Plus Enzymatic Chromatin IP Kit (Agarose Beads) #9004
Note: Agarose beads are blocked with sonicated salmon sperm DNA, which will contaminate DNA library preps and NG-seq.
Required Reagents:
Reagents Included:
Reagents Not Included:
Illumina Systems NG-seq platforms use a red laser/LED to sequence A/C and a green laser/LED to sequence G/T. For each cycle, both the red and the green channel need to be read to ensure proper image registration (i.e. A or C must be present in each cycle, and G or T must be present in each cycle). If this color balance is not maintained, sequencing the index read could fail. Please check the sequences of each index to be used to ensure that you will have signal in both the red and green channels for every cycle. See example below:
| Index 7 Primers for Illumina Systems | Index 5 Primers for Illumina Systems | ||
| Index 1 Primer for Illumina Systems |
ATCACG |
Index 1 Primer for Illumina Systems |
ATCACG |
| Index 2 Primer for Illumina Systems |
CGATGT |
Index 2 Primer for Illumina Systems |
CGATGT |
| Index 4 Primer for Illumina Systems |
TGACCA |
Index 3 Primer for Illumina Systems |
TTAGGC |
| Index 7 Primer for Illumina Systems |
CAGATC |
Index 4 Primer for Illumina Systems |
TGACCA |
| ✔✔✔✔✔✔ | ✔✘✘✔✔✔ | ||
| Pool of 2 samples | Index 6 and 12 Primers |
| Pool of 3 samples | Index 4, 6, and 12 Primers |
| Pool of 6 samples | Index 2, 4, 5, 6, 7, and 12 Primers |
Multiplex Oligos for Illumina Systems (Single Index Primers) (ChIP-seq, CUT&RUN) can generate 12 different, barcoded samples if each Index primer is used only once with Universal PCR Primer for Illumina Systems. Each Index Primer is supplied in sufficient amounts to generate two libraries, but these two libraries cannot be pooled together in one sequencing lane.
For 1-plex (no pooling), use any index primer with the universal PCR primer.
Each Index Primer for Illumina Systems is provided in volume of 10 µl.
| Index 1 Primer for Illumina Systems | 5´-CAAGCAGAAGACGGCATACGAGATCGTGATGTGACTGGAG- TTCAGACGTGTGCTCTTCCGATC-s-T-3´ |
ATCACG |
| Index 2 Primer for Illumina Systems | 5´-CAAGCAGAAGACGGCATACGAGATACATCGGTGACTGGAG- TTCAGACGTGTGCTCTTCCGATC-s-T-3´ |
CGATGT |
| Index 3 Primer for Illumina Systems | 5´-CAAGCAGAAGACGGCATACGAGATGCCTAAGTGACTGGA- GTTCAGACGTGTGCTCTTCCGATC-s-T-3´ |
TTAGGC |
| Index 4 Primer for Illumina Systems | 5´-CAAGCAGAAGACGGCATACGAGATTGGTCAGTGACTGGA- GTTCAGACGTGTGCTCTTCCGATC-s-T-3´ |
TGACCA |
| Index 5 Primer for Illumina Systems | 5´-CAAGCAGAAGACGGCATACGAGATCACTGTGTGACTGGA- GTTCAGACGTGTGCTCTTCCGATC-s-T-3´ |
ACAGTG |
| Index 6 Primer for Illumina Systems | 5´-CAAGCAGAAGACGGCATACGAGATATTGGCGTGACTGGAG- TTCAGACGTGTGCTCTTCCGATC-s-T-3´ |
GCCAAT |
| Index 7 Primer for Illumina Systems | 5´-CAAGCAGAAGACGGCATACGAGATGATCTGGTGACTGGAG- TTCAGACGTGTGCTCTTCCGATC-s-T-3´ |
CAGATC |
| Index 8 Primer for Illumina Systems | 5´-CAAGCAGAAGACGGCATACGAGATTCAAGTGTGACTGGAG- TTCAGACGTGTGCTCTTCCGATC-s-T-3´ |
ACTTGA |
| Index 9 Primer for Illumina Systems | 5´-CAAGCAGAAGACGGCATACGAGATCTGATCGTGACTGGAG- TTCAGACGTGTGCTCTTCCGATC-s-T-3´ |
GATCAG |
| Index 10 Primer for Illumina Systems | 5´-CAAGCAGAAGACGGCATACGAGATAAGCTAGTGACTGGAG- TTCAGACGTGTGCTCTTCCGATC-s-T-3´ |
TAGCTT |
| Index 11 Primer for Illumina Systems | 5´-CAAGCAGAAGACGGCATACGAGATGTAGCCGTGACTGGA- TTCAGACGTGTGCTCTTCCGATC-s-T-3´ |
GGCTAC |
| Index 12 Primer for Illumina Systems | 5´-CAAGCAGAAGACGGCATACGAGATTACAAGGTGACTGGAG- TTCAGACGTGTGCTCTTCCGATC-s-T-3´ |
CTTGTA |
Where -s- indicates phosphorothioate bond.
The components in the SimpleChIP® ChIP-seq Multiplex Oligos for Illumina Systems (Single Index Primers) #29580 are individually validated by the functional testing listed below and must pass rigorous quality control standards. Furthermore, each set of components is functionally validated together by construction and sequencing of indexed libraries on the Illumina Systems sequencing platform.
5´-/5Phos/GAT CGG AAG AGC ACA CGT CTG AAC TCC AGT C/ideoxyU/A CAC TCT TTC CCT ACA CGA CGC TCT TCC GAT C-s-T-3´
Quality Control Assays
Supplied in: 50 mM KCl, 5 mM NaCl, 10 mM Tris-HCl (pH 7.4 @ 25°C), 0.1 mM EDTA, 1 mM DTT, 175 μg/ml BSA and 50% Glycerol
Quality Control Assays
5´-AAT GAT ACG GCG ACC ACC GAG ATC TAC ACT CTT TCC CTA CAC GAC GCT CTT CCG ATC*T-3´
Quality Control Assays
Quality Control Assays
posted November 2017
revised January 2023
protocol id: 1626
Protocol Id: 1626
Next generation sequencing (NG-seq) is a high throughput method that can be used downstream of chromatin immunoprecipitation (ChIP) and Cleavage Under Targets and Release Using Nuclease (CUT&RUN) assays to identify and quantify target DNA enrichment across the entire genome. Multiplex Oligos for Illumina® (Dual Index Primers) (ChIP-seq, CUT&RUN) contains adaptors and primers that are ideally suited for multiplex sample preparation for NG-seq on the Illumina® platform (Illumina, Inc.). This kit can be used to generate up to 96 distinct, barcoded ChIP-seq or CUT&RUN DNA libraries that can be combined into a single sequencing reaction.
Each kit component must pass rigorous quality control standards, and for each new lot the entire set of reagents is functionally validated together by construction and sequencing of indexed libraries on the Illumina® sequencing platform.
This product provides enough reagents to support up to 96 DNA sequencing libraries, and must be used in combination with DNA Library Prep Kit for Illumina® (ChIP-seq, CUT&RUN) #56795.
Compatible Assay kits:
SimpleChIP® Enzymatic Chromatin IP Kit (Magnetic Beads) #9003
SimpleChIP® Plus Enzymatic Chromatin IP Kit (Magnetic Beads) #9005
SimpleChIP® Plus Sonication Chromatin IP Kit #56383
ChIP-seq DNA Library Prep Kit for Illumina® (ChIP-seq, CUT&RUN) #56795
CUT&RUN Assay Kit #86652
Non-Compatible SimpleChIP® kits:
SimpleChIP® Enzymatic Chromatin IP Kit (Agarose Beads) #9002
SimpleChIP® Plus Enzymatic Chromatin IP Kit (Agarose Beads) #9004
Note: Agarose beads are blocked with sonicated salmon sperm DNA, which will contaminate DNA library preps and NG-seq.
Required Reagents:
Reagents Included:
Reagents Not Included:
The dual index primer strategy utilizes two 8 base indices within each primer. Index 7 primers contain indices that are adjacent to the P7 sequence while index 5 primers contain indices that are adjacent to the P5 sequence. Dual indexing is enabled by adding a unique index to both ends of a sample to be sequenced. Up to 96 different samples can be uniquely indexed by combining each of the 12 index 7 primers with each of the 8 index 5 primers.
Illumina® NG-seq platforms use a red laser/LED to sequence A/C and a green laser/LED to sequence G/T. For each cycle, both the red and the green channel need to be read to ensure proper image registration (i.e. A or C must be present in each cycle, and G or T must be present in each cycle). If this color balance is not maintained, sequencing the index read could fail. Please check the sequences of each index to be used to ensure that you will have signal in both the red and green channels for every cycle. See example below:
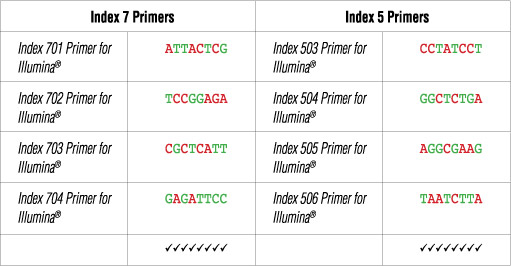
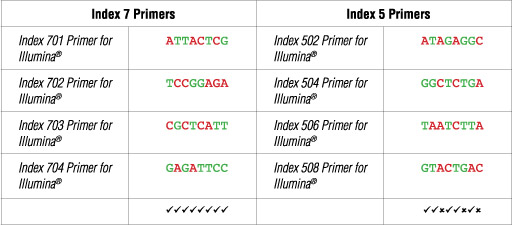
The following table lists some (but not all) valid index combinations that can be sequenced together:

Some other valid combinations are listed below. Choose a valid set of Index 7 primers and a valid set of Index 5 primers. Use each Index 7 primer with each i5 primer to form desired number of primer pairs for PCR amplification of desired number of libraries.

Each Index 5 Primer for Illumina® is provided in volume of 60 µl.

Where -s- indicates phosphorothioate bond.
Each Index 7 Primer for Illumina® is provided in a volume of 40 µl.
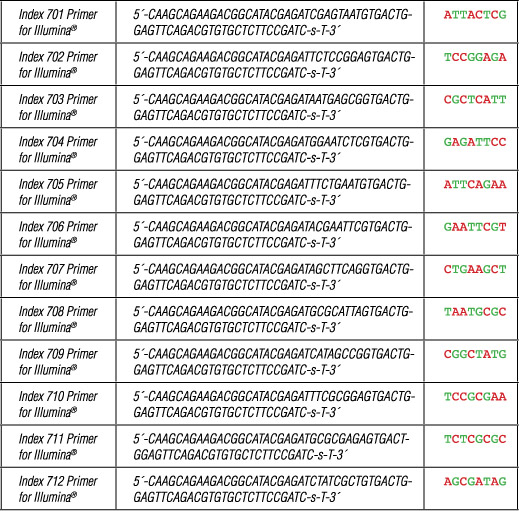
Where -s- indicates phosphorothioate bond.
The components in the Multiplex Oligos for Illumina® (Dual Index Primers) (ChIP-seq, CUT&RUN) #47538 are individually validated by the functional testing listed below and must pass rigorous quality control standards. Furthermore, each set of components is functionally validated together by construction and sequencing of indexed libraries on the Illumina® sequencing platform.
5´-/5Phos/GAT CGG AAG AGC ACA CGT CTG AAC TCC AGT C/ideoxyU/A CAC TCT TTC CCT ACA CGA CGC TCT TCC GAT C-s-T-3´
Quality Control Assays
Supplied in: 50 mM KCl, 5 mM NaCl, 10 mM Tris-HCl (pH 7.4 @ 25°C), 0.1 mM EDTA, 1 mM DTT, 175 µg/ml BSA, and 50% Glycerol
Quality Control Assays
Quality Control Assays
posted November 2017
revised April 2022
protocol id: 1625
Protocol Id: 2806
The chromatin immunoprecipitation (ChIP) assay is a powerful and versatile technique used for probing protein-DNA interactions within the natural chromatin context of the cell (1,2). This assay can be used to identify multiple proteins associated with a specific region of the genome, or the opposite, to identify the many regions of the genome bound by a particular protein (3-6). It can be used to determine the specific order of recruitment of various proteins to a gene promoter or to "measure" the relative amount of a particular histone modification across an entire gene locus (3,4). In addition to histone proteins, the ChIP assay can be used to analyze binding of transcription factors and co-factors, DNA replication factors and DNA repair proteins. When performing the ChIP assay, cells or tissues are first fixed with formaldehyde, a reversible protein-DNA cross-linking agent that "preserves" the protein-DNA interactions occurring in the cell (1,2). Cells are lysed and chromatin is harvested and fragmented using either sonication or enzymatic digestion. The chromatin is then immunoprecipitated with antibodies specific to a particular protein or histone modification. Any DNA sequences that are associated with the protein or histone modification of interest will co-precipitate as part of the cross-linked chromatin complex and the relative amount of that DNA sequence will be enriched by the immunoselection process. After immunoprecipitation, the protein-DNA cross-links are reversed and the DNA is purified. Standard PCR or Quantitative Real-Time PCR can be used to measure the amount of enrichment of a particular DNA sequence by a protein-specific immunoprecipitation (1,2). Alternatively, the ChIP assay can be combined with genomic tiling micro-array (ChIP on chip) techniques, high throughput sequencing, or cloning strategies, all of which allow for genome-wide analysis of protein-DNA interactions and histone modifications (5-8).
Except as otherwise expressly agreed in a writing signed by a legally authorized representative of CST, the following terms apply to Products provided by CST, its affiliates or its distributors. Any Customer's terms and conditions that are in addition to, or different from, those contained herein, unless separately accepted in writing by a legally authorized representative of CST, are rejected and are of no force or effect.
Products are labeled with For Research Use Only or a similar labeling statement and have not been approved, cleared, or licensed by the FDA or other regulatory foreign or domestic entity, for any purpose. Customer shall not use any Product for any diagnostic or therapeutic purpose, or otherwise in any manner that conflicts with its labeling statement. Products sold or licensed by CST are provided for Customer as the end-user and solely for research and development uses. Any use of Product for diagnostic, prophylactic or therapeutic purposes, or any purchase of Product for resale (alone or as a component) or other commercial purpose, requires a separate license from CST. Customer shall (a) not sell, license, loan, donate or otherwise transfer or make available any Product to any third party, whether alone or in combination with other materials, or use the Products to manufacture any commercial products, (b) not copy, modify, reverse engineer, decompile, disassemble or otherwise attempt to discover the underlying structure or technology of the Products, or use the Products for the purpose of developing any products or services that would compete with CST products or services, (c) not alter or remove from the Products any trademarks, trade names, logos, patent or copyright notices or markings, (d) use the Products solely in accordance with CST Product Terms of Sale and any applicable documentation, and (e) comply with any license, terms of service or similar agreement with respect to any third party products or services used by Customer in connection with the Products.