Revision 11
#56795
Store at -20C
877-616-CELL (2355)
877-678-TECH (8324)
3 Trask Lane | Danvers | Massachusetts | 01923 | USA
For Research Use Only. Not for Use in Diagnostic Procedures.
Applications:
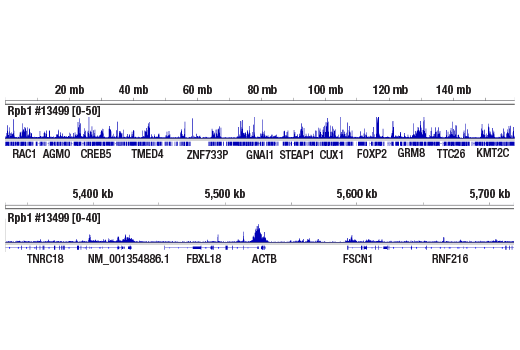
ChIP-seq, C&R
Reactivity:
All
Storage
Specificity/Sensitivity
Description
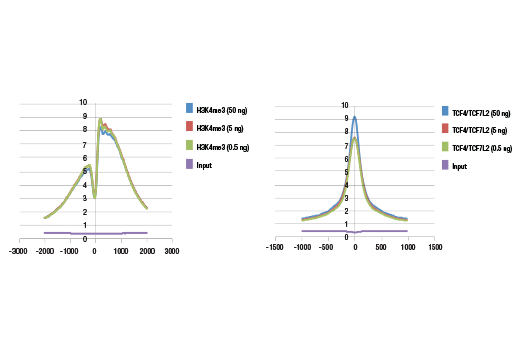
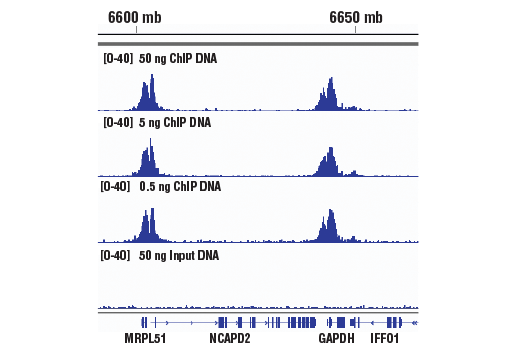
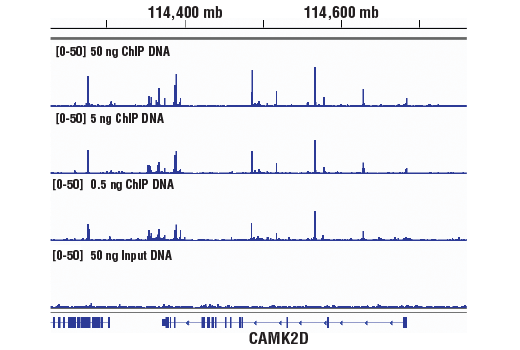
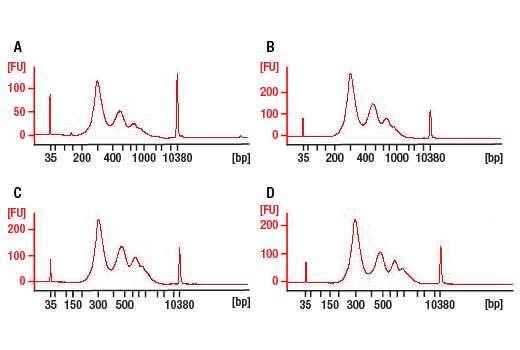
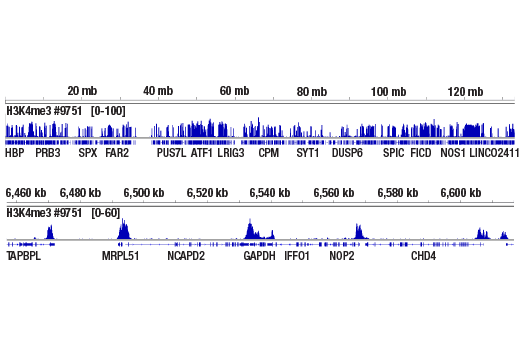
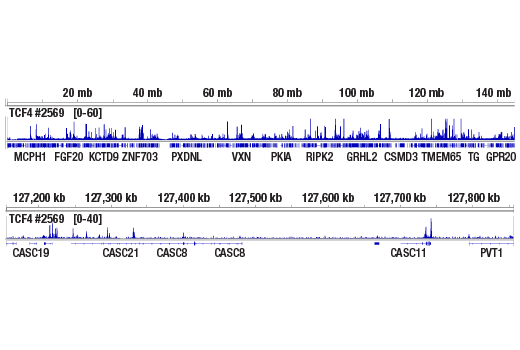
This product provides sufficient amounts of reagents for 24 reactions and is compatible with both enzymatic- or sonication-fragmented, ChIP-enriched DNA and CUT&RUN DNA. Distinct protocols are provied for DNA library preparation from ChIP and CUT&RUN DNA. This product is compatible with SimpleChIP® Enzymatic Chromatin IP Kit (Magnetic Beads) #9003, SimpleChIP® Plus Enzymatic Chromatin IP Kit (Magnetic Beads) #9005, SimpleChIP® Plus Sonication Chromatin IP Kit #56383, CUT&RUN Assay Kit #86652, and CUT&RUN Assay Kit (w/ Drosophila Spike-In Control) #84647. This product is not compatible with CUT&Tag Assay Kit #77552, SimpleChIP® Enzymatic Chromatin IP Kit (Agarose Beads) #9002 and SimpleChIP® Plus Enzymatic Chromatin IP Kit (Agarose Beads) #9004 because agarose beads are blocked with sonicated salmon sperm DNA, which will contaminate DNA library preps and NG-seq.
Background
Background References
- Orlando, V. (2000) Trends Biochem Sci 25, 99-104.
- Kuo, M.H. and Allis, C.D. (1999) Methods 19, 425-33.
- Agalioti, T. et al. (2000) Cell 103, 667-78.
- Soutoglou, E. and Talianidis, I. (2002) Science 295, 1901-4.
- Mikkelsen, T.S. et al. (2007) Nature 448, 553-60.
- Lee, T.I. et al. (2006) Cell 125, 301-13.
- Weinmann, A.S. and Farnham, P.J. (2002) Methods 26, 37-47.
- Wells, J. and Farnham, P.J. (2002) Methods 26, 48-56.
Species Reactivity
Species reactivity is determined by testing in at least one approved application (e.g., western blot).
Applications Key
ChIP-seq: Chromatin IP-seq C&R: CUT&RUN
Cross-Reactivity Key
All: All Species Expected
Trademarks and Patents
Cell Signaling Technology is a trademark of Cell Signaling Technology, Inc.
SimpleChIP is a registered trademark of Cell Signaling Technology, Inc.
All other trademarks are the property of their respective owners. Visit cellsignal.com/trademarks for more information.
Limited Uses
Except as otherwise expressly agreed in a writing signed by a legally authorized representative of CST, the following terms apply to Products provided by CST, its affiliates or its distributors. Any Customer's terms and conditions that are in addition to, or different from, those contained herein, unless separately accepted in writing by a legally authorized representative of CST, are rejected and are of no force or effect.
Products are labeled with For Research Use Only or a similar labeling statement and have not been approved, cleared, or licensed by the FDA or other regulatory foreign or domestic entity, for any purpose. Customer shall not use any Product for any diagnostic or therapeutic purpose, or otherwise in any manner that conflicts with its labeling statement. Products sold or licensed by CST are provided for Customer as the end-user and solely for research and development uses. Any use of Product for diagnostic, prophylactic or therapeutic purposes, or any purchase of Product for resale (alone or as a component) or other commercial purpose, requires a separate license from CST. Customer shall (a) not sell, license, loan, donate or otherwise transfer or make available any Product to any third party, whether alone or in combination with other materials, or use the Products to manufacture any commercial products, (b) not copy, modify, reverse engineer, decompile, disassemble or otherwise attempt to discover the underlying structure or technology of the Products, or use the Products for the purpose of developing any products or services that would compete with CST products or services, (c) not alter or remove from the Products any trademarks, trade names, logos, patent or copyright notices or markings, (d) use the Products solely in accordance with CST Product Terms of Sale and any applicable documentation, and (e) comply with any license, terms of service or similar agreement with respect to any third party products or services used by Customer in connection with the Products.
Revision 11
Revision 11
Revision 11