Revision 6
#40366
Store at -20C
877-616-CELL (2355)
877-678-TECH (8324)
3 Trask Lane | Danvers | Massachusetts | 01923 | USA
For Research Use Only. Not for Use in Diagnostic Procedures.
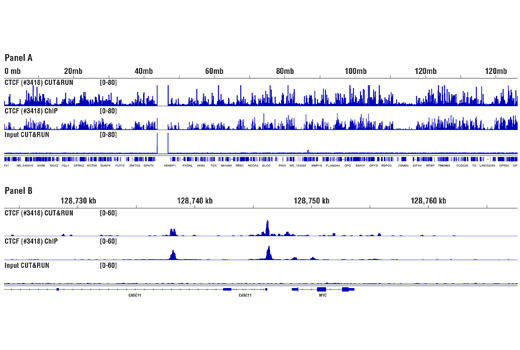
Applications:
C&R
Product Usage Information
For the pAG-MNase Enzyme, after cell permeabilization and primary antibody binding, resuspend cells in 50 μl of digitonin buffer containing 1.5 μl of pAG-MNase Enzyme (33X dilution). Incubate cell samples with rotation at 4°C for 1 hour, wash cells with digitonin buffer, and then perform the chromatin digestion.
Sample Normalization Spike-In DNA can be added directly to the digestion stop buffer. For sample normalization with NG-seq, add 5 μl (50 pg) of Sample Normalization Spike-In DNA
to each reaction. When using 100,000 cells or 1mg of tissue per reaction this ensures that the normalization reads are around 0.5% of the total sequencing reads. If more or less than 100,000 cells or 1mg of tissue are used per reaction, proportionally scale the volume of Sample Normalization Spike-In DNA up or down to adjust normalization reads to around 0.5% of total reads.
When performing sample normalization, be sure to map the CUT&RUN sequencing data for all samples to both the test reference genome (e.g. human) and the sample normalization genome (S. cerevisiae).
Storage
Description
Background
Species Reactivity
Species reactivity is determined by testing in at least one approved application (e.g., western blot).
Applications Key
C&R: CUT&RUN
Trademarks and Patents
Cell Signaling Technology is a trademark of Cell Signaling Technology, Inc.
SimpleChIP is a registered trademark of Cell Signaling Technology, Inc.
All other trademarks are the property of their respective owners. Visit cellsignal.com/trademarks for more information.
Limited Uses
Except as otherwise expressly agreed in a writing signed by a legally authorized representative of CST, the following terms apply to Products provided by CST, its affiliates or its distributors. Any Customer's terms and conditions that are in addition to, or different from, those contained herein, unless separately accepted in writing by a legally authorized representative of CST, are rejected and are of no force or effect.
Products are labeled with For Research Use Only or a similar labeling statement and have not been approved, cleared, or licensed by the FDA or other regulatory foreign or domestic entity, for any purpose. Customer shall not use any Product for any diagnostic or therapeutic purpose, or otherwise in any manner that conflicts with its labeling statement. Products sold or licensed by CST are provided for Customer as the end-user and solely for research and development uses. Any use of Product for diagnostic, prophylactic or therapeutic purposes, or any purchase of Product for resale (alone or as a component) or other commercial purpose, requires a separate license from CST. Customer shall (a) not sell, license, loan, donate or otherwise transfer or make available any Product to any third party, whether alone or in combination with other materials, or use the Products to manufacture any commercial products, (b) not copy, modify, reverse engineer, decompile, disassemble or otherwise attempt to discover the underlying structure or technology of the Products, or use the Products for the purpose of developing any products or services that would compete with CST products or services, (c) not alter or remove from the Products any trademarks, trade names, logos, patent or copyright notices or markings, (d) use the Products solely in accordance with CST Product Terms of Sale and any applicable documentation, and (e) comply with any license, terms of service or similar agreement with respect to any third party products or services used by Customer in connection with the Products.
Revision 6
Revision 6