CUT&RUN Overview
Learn about CUT&RUN - a low cell number alternative to ChIP-qPCR and ChIP-seq to analyze protein-DNA interaction in chromatin.
What is CUT&RUN?
The development of methods that enable mapping of protein-DNA interactions, such as chromatin-immunoprecipitation (ChIP) and ChIP-seq, have led to a growing awareness that aberrant epigenetic regulation drives a wide variety of human diseases. Cleavage Under Targets & Release Using Nuclease (CUT&RUN) is a new technology that can be used for chromatin profiling.
How It Works
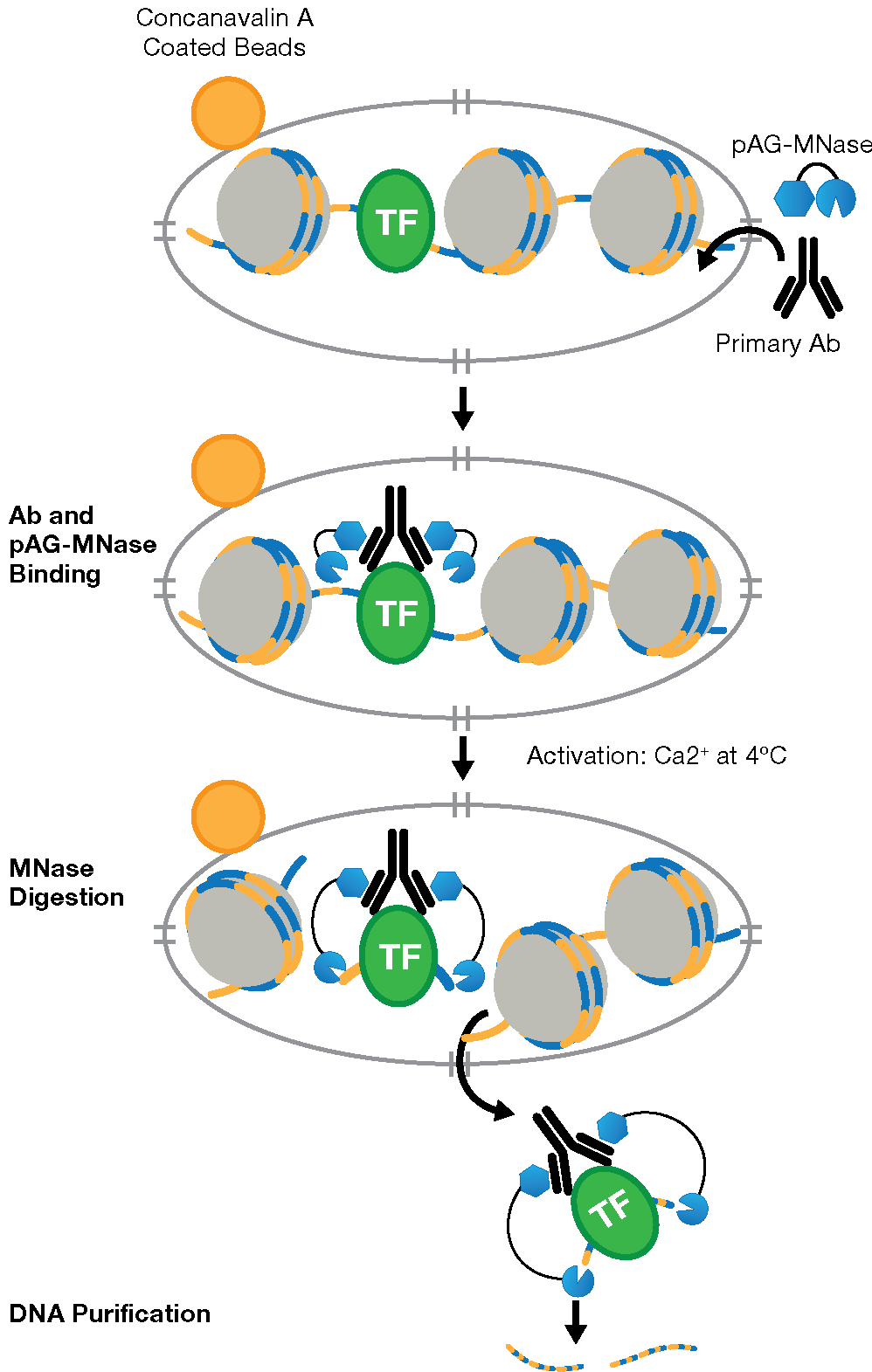
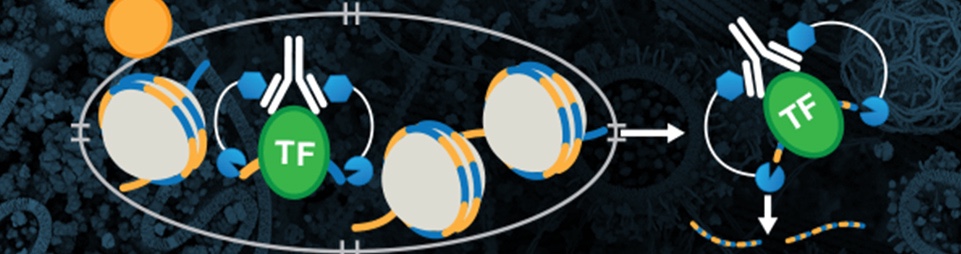
CUT&RUN is an in vivo method that uses a target-specific primary antibody and a Protein A-Protein G-Micrococcal Nuclease (pAG-MNase) to isolate specific protein-DNA complexes1,2,3. It only takes 1 to 2 days to get from cell to DNA and can be automated for maximal throughput and reproducibility4.
To isolate the protein-DNA complex of interest, cells are first harvested and bound to Concanavalin A-coated magnetic beads to simplify cell handling and minimize cell loss during subsequent washes. Cell membranes are permeabilized with digitonin to facilitate the entry of the primary antibody into the nuclei, where it binds to the histone, transcription factor, or cofactor of interest. The pAG domain of the pAG-MNase fusion protein then binds to the primary antibody heavy chain, targeting the enzyme to the chromatin region of interest. The addition of Ca2+ activates the pAG-MNase to initiate DNA digestion. This allows the cleaved chromatin complex to diffuse away from the genomic chromatin, out of the nuclei, and into the sample supernatant, where it can be collected using either a DNA spin column or phenol/chloroform extraction followed by ethanol precipitation.
The purified, enriched DNA can then be identified and quantified using qPCR or used to construct a DNA sequencing library for next-generation sequencing (NGS) and whole-genome mapping.
CUT&RUN Benefits
Cell Signaling Technology® (CST) offers a CUT&RUN kit that overcomes many of the challenges faced with other whole-genome mapping techniques. Benefits offered by the CST CUT&RUN kit include:
Fast time to results | 1 to 2 days from cell to DNA |
Low sample requirement | 100K cells recommended, validated for 5-20K cells if needed (dependent on target type) |
Sequencing cost savings | Only 3 to 5 million high-quality reads required |
Avoid “crosslinking” artifacts | An in vivo method performed using native chromatin |
Antibody versatility | Compatible with rabbit and mouse antibodies |
Target versatility | Generate sequencing and/or qPCR data for histones, histone modifications, transcription factors, and cofactors |
Reproducible results | Spike in control DNA to normalize signal between samples |
Low Sample Requirement
Save time by only culturing 100K cells to look at protein-DNA interactions and/or get epigenetic information for low abundance cell types.
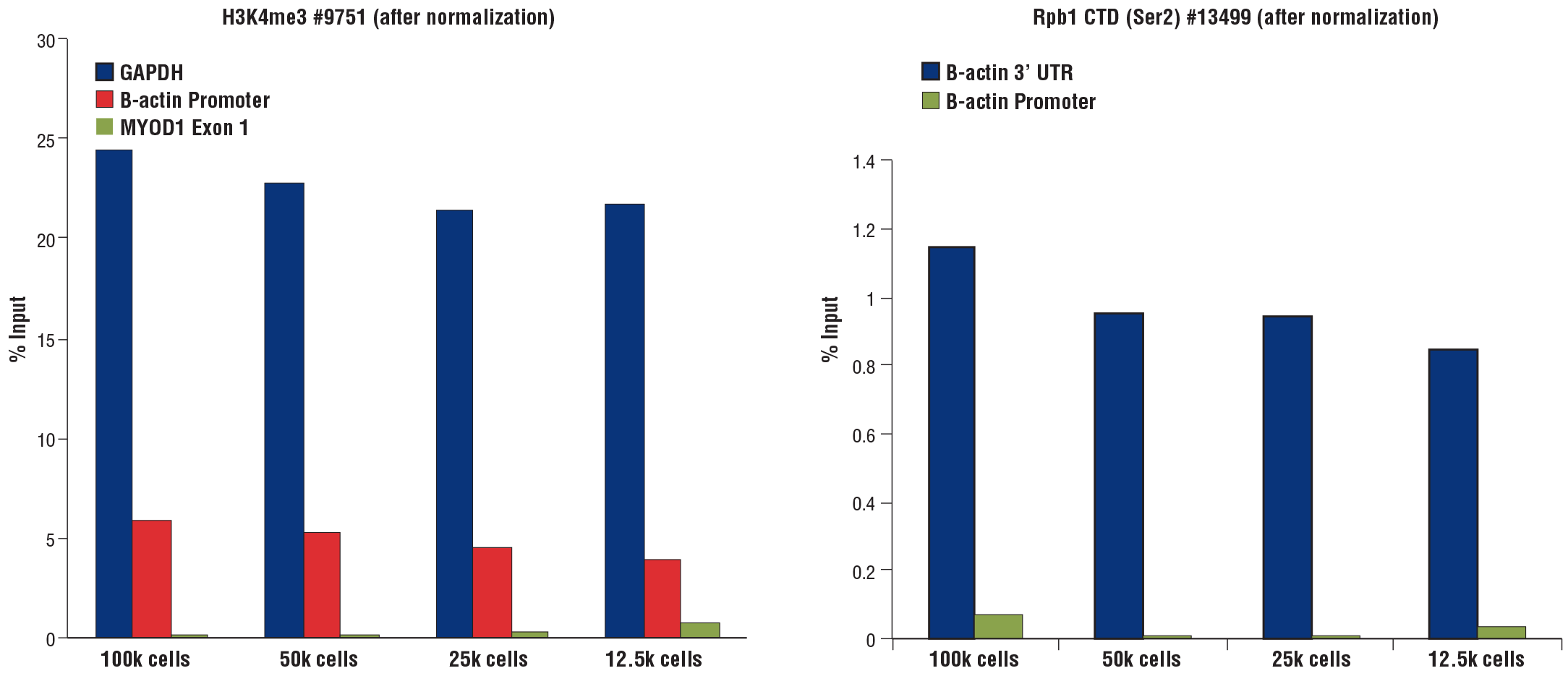
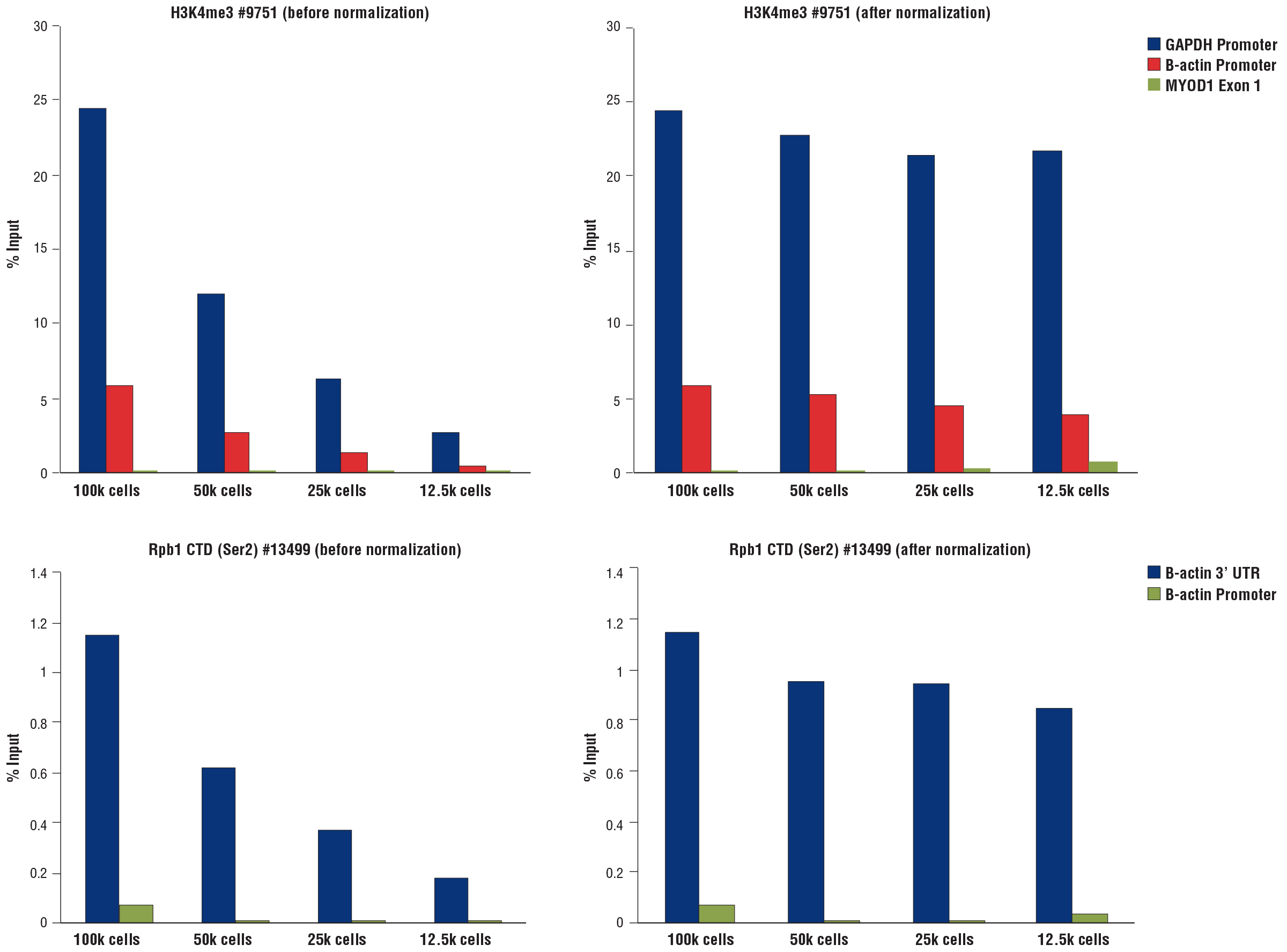
Similar results were obtained when decreasing numbers of HCT 116 cells were used to perform CUT&RUN using the CUT&RUN Assay Kit #86652. Assays were performed with either Tri-Methyl-Histone H3 (Lys4) (C42D8) Rabbit mAb #9751 (left panel) or Phospho-Rpb1 CTD (Ser2) (E1Z3G) Rabbit mAb #13499 (right panel). Enriched DNA was quantified by real-time PCR using the SimpleChIP® Universal qPCR Master Mix #88989 and SimpleChIP® Human GAPDH Exon 1 Primers, SimpleChIP® Human β-Actin Promoter Primers #13653, SimpleChIP® Human β-Actin 3' UTR Primers #13669, and SimpleChIP® Human MyoD1 Exon 1 Primers #4490. The amount of immunoprecipitated DNA in each sample is represented as the signal relative to the total amount of input chromatin (percent input for 100,000 cells). The data are normalized using spike-in DNA added to each reaction.
Fast Time to Results
Save on the time it takes to enrich for the target chromatin. Spend just 1 to 2 days to get from cells to DNA.
Sequencing Cost Savings
The relatively low background means you can distinguish your signal from genomic background noise with a lower sequencing depth. Sequencing data can be obtained with just 3 to 5 million high-quality reads, reducing your sequencing costs significantly. An added bonus? The method’s low background also makes it possible to profile low-signal genomic features.
Target Versatility
Purified, enriched DNA can be used for sequencing and/or qPCR experiments to investigate histones, histone modifications, transcription factors, and cofactors.
Histone Modifications
CUT&RUN Assay Kit #86652 works as well as ChIP-qPCR and ChIP-seq when studying histone modifications and only requires 100,000 cells.
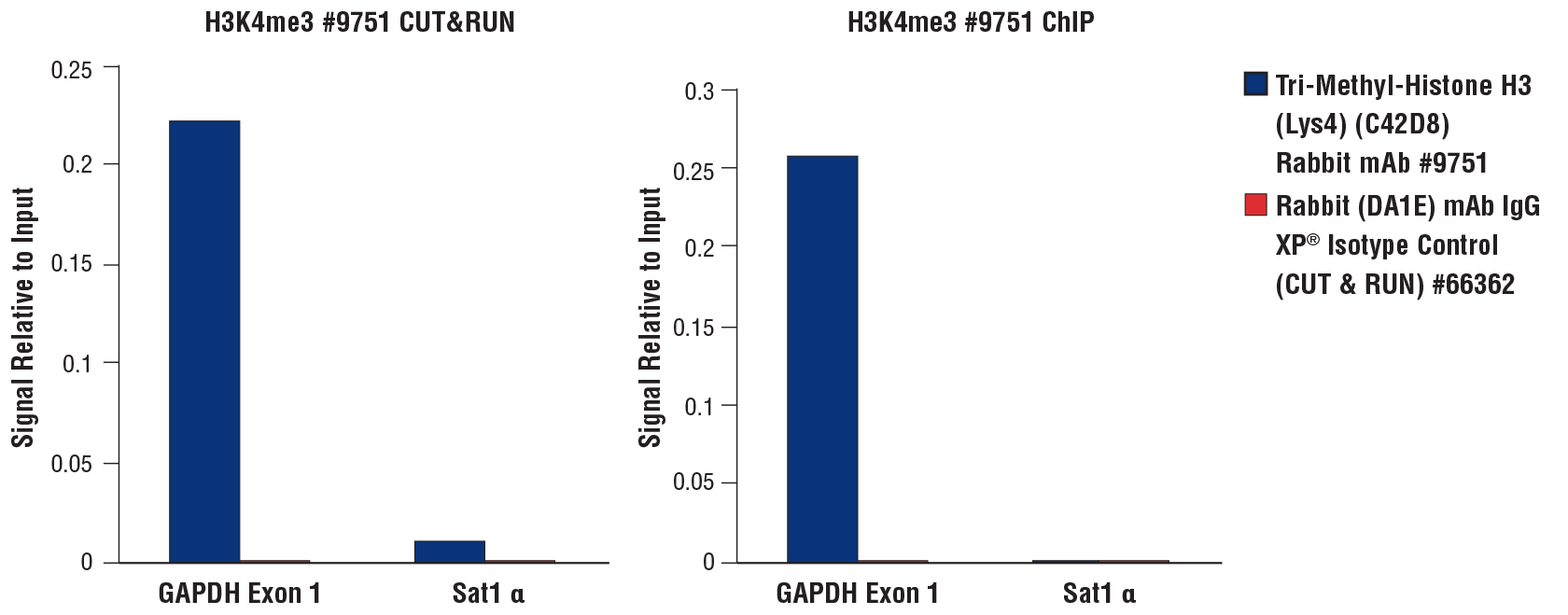
CUT&RUN and ChIP assays were performed with HCT 116 cells (1x105 cells for CUT&RUN, 4x106 cells for ChIP) and Tri-Methyl-Histone H3 (Lys4) (C42D8) Rabbit mAb #9751 or Rabbit (DA1E) mAb IgG XP® Isotype Control (CUT&RUN) #66362, using the CUT&RUN Assay Kit #86652 (left panel) or the SimpleChIP® Plus Enzymatic Chromatin IP Kit (Magnetic Beads) #9005 (right panel). The enriched DNA was quantified by real-time PCR using the SimpleChIP® Universal qPCR Master Mix #88989, and SimpleChIP® Human GAPDH Exon 1 Primers, and SimpleChIP® Human α Satellite Repeat Primers #4486. The amount of immunoprecipitated DNA in each sample is represented as the signal relative to the total amount of input chromatin, which is equivalent to 1.
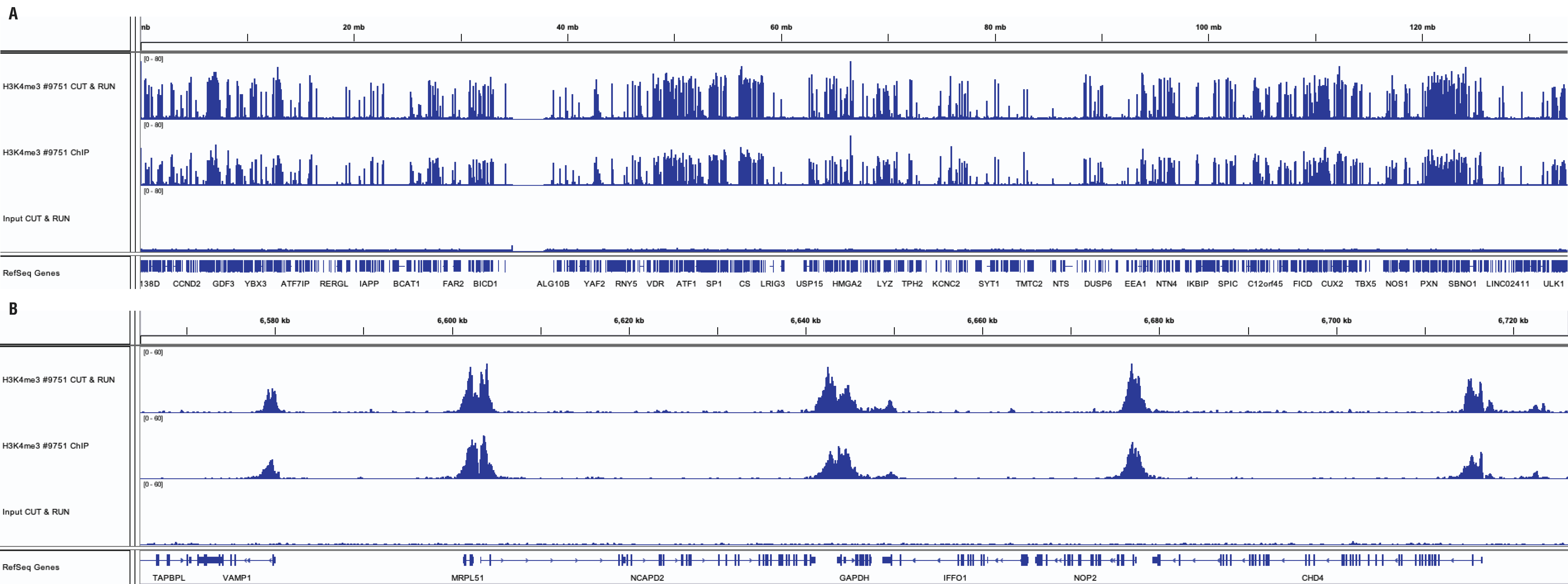
CUT&RUN and ChIP assays were performed with HCT 116 cells (1x105 cells for CUT&RUN, 4x106 cells for ChIP) and Tri-Methyl-Histone H3 (Lys4) (C42D8) Rabbit mAb #9751, using the CUT&RUN Assay Kit #86652 or the SimpleChIP® Plus Enzymatic Chromatin IP Kit (Magnetic Beads) #9005. DNA libraries were prepared using DNA Library Prep Kit for Illumina (ChIP-seq, CUT&RUN) #56795. Panel A compares enrichment of H3K4me3 across chromosome 12, while Panel B compares enrichment at the GAPDH gene, a known target of H3K4me3. The input tracks are from the CUT&RUN input sample.
Transcription Factors
CUT&RUN Assay Kit #86652 works as well as ChIP-qPCR and ChIP-seq when studying transcription factor DNA-protein interactions and only requires 100,000 cells.
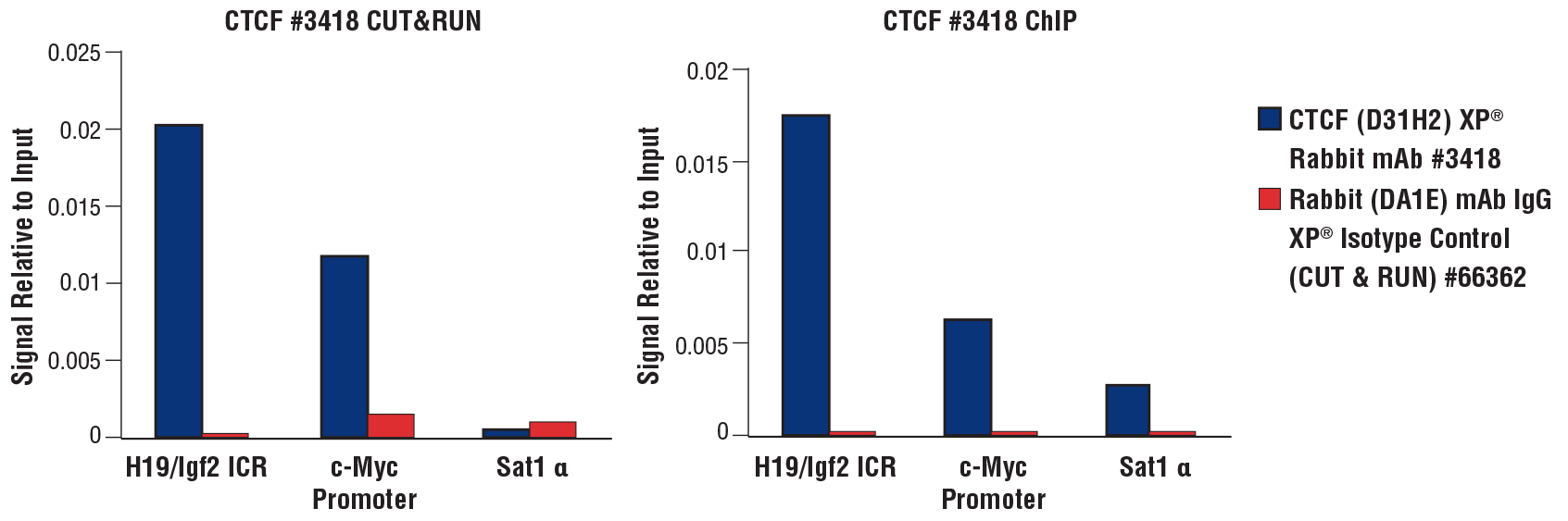
CUT&RUN and ChIP assays were performed with HCT 116 cells (1x105 cells for CUT&RUN, 4x106 cells for ChIP) and CTCF (D31H2) XP® Rabbit mAb #3418 or Rabbit (DA1E) mAb IgG XP® Isotype Control (CUT&RUN) #66362, using the CUT&RUN Assay Kit #86652 (left panel) or the SimpleChIP® Plus Enzymatic Chromatin IP Kit (Magnetic Beads) #9005 (right panel). The enriched DNA was quantified by real-time PCR using the SimpleChIP® Universal qPCR Master Mix #88989 and human c-Myc promoter primers, SimpleChIP® Human H19/Igf2 ICR Primers, and SimpleChIP® Human α Satellite Repeat Primers #4486. The amount of immunoprecipitated DNA in each sample is represented as the signal relative to the total amount of input chromatin, which is equivalent to 1.
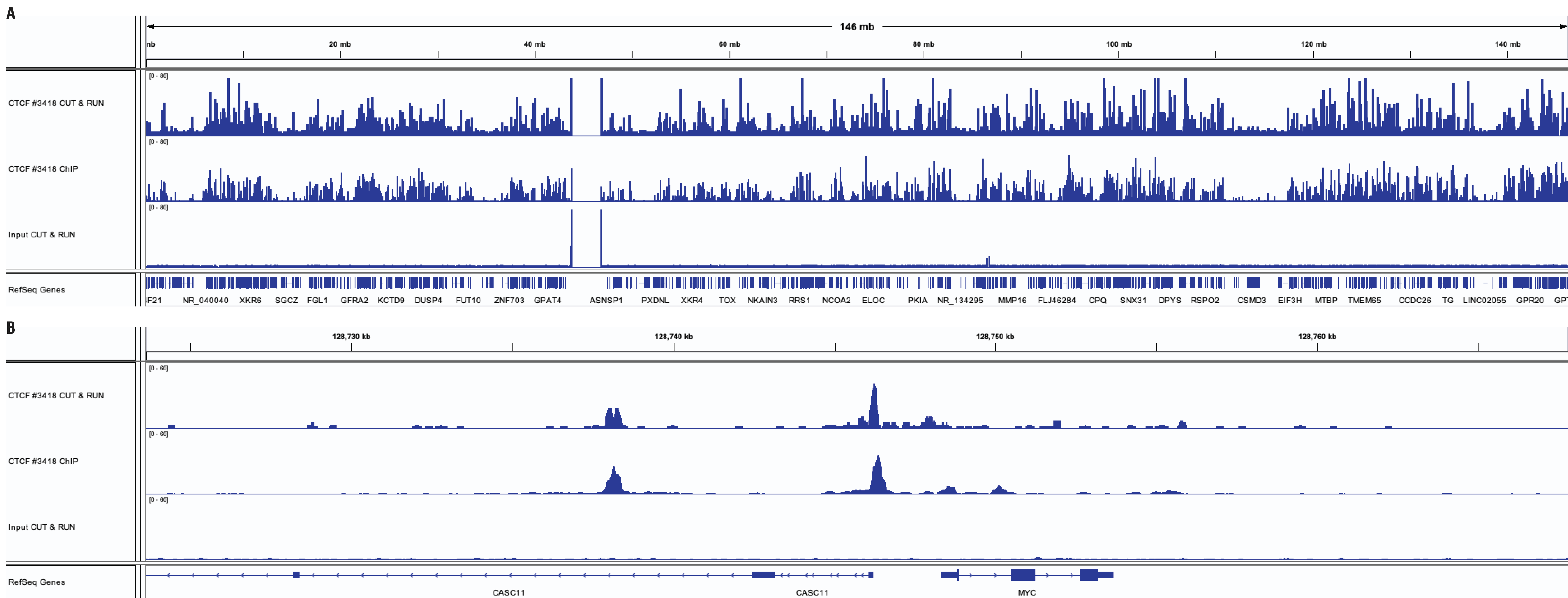
CUT&RUN and ChIP assays were performed with HCT 116 cells (1x105 cells for CUT&RUN, 4x106 cells for ChIP) and CTCF (D31H2) XP® Rabbit mAb #3418, using the CUT&RUN Assay Kit #86652 or the SimpleChIP® Plus Enzymatic Chromatin IP Kit (Magnetic Beads) #9005. DNA libraries were prepared using DNA Library Prep Kit for Illumina (ChIP-seq, CUT&RUN) #56795. Panel A compares enrichment of CTCF across chromosome 8, while Panel B compares enrichment at the MYC gene, a known target of CTCF. The input tracks are from the CUT&RUN input sample.
Cofactors
CUT&RUN Assay Kit #86652 works as well as ChIP-qPCR and ChIP-seq when studying cofactor DNA-protein interactions and only requires 100,000 cells.
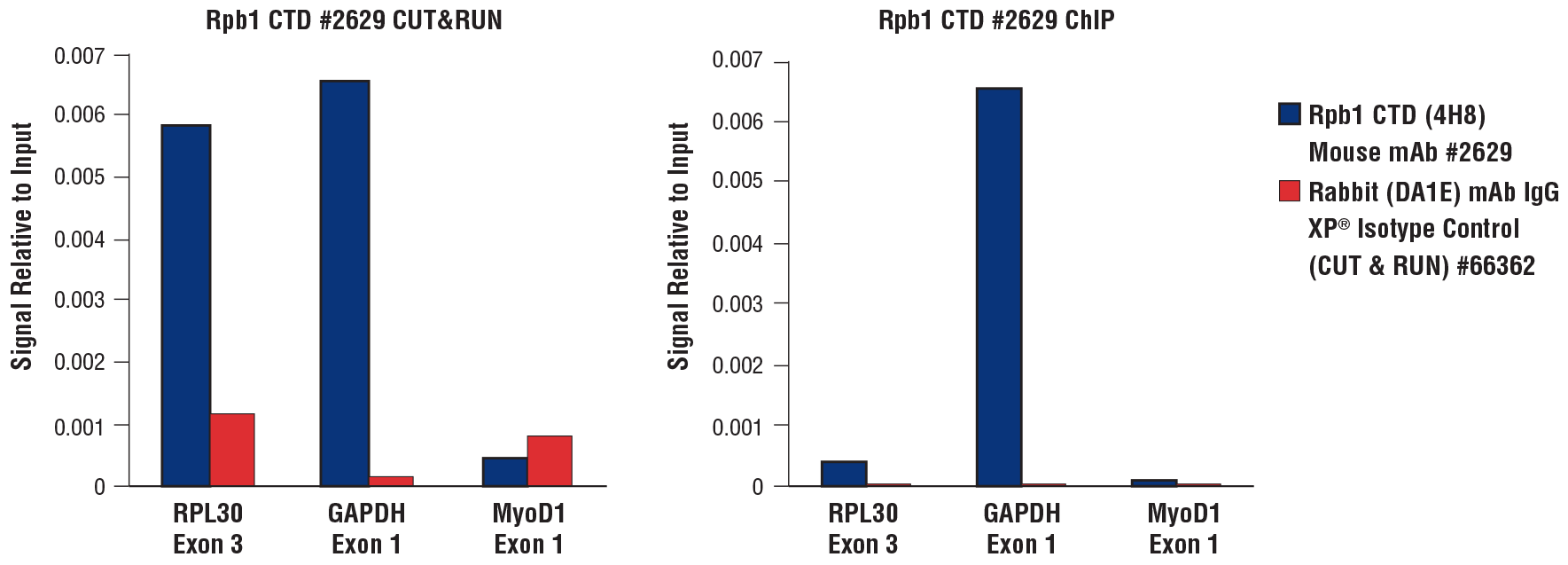
CUT&RUN and ChIP assays were performed with HeLa cells (1x105 cells for CUT&RUN, 4x106 cells for ChIP) and Rpb1 CTD (4H8) Mouse mAb #2629 or Rabbit (DA1E) mAb IgG XP® Isotype Control (CUT&RUN) #66362, using the CUT&RUN Assay Kit #86652 (left panel) or the SimpleChIP® Plus Enzymatic Chromatin IP Kit (Magnetic Beads) #9005 (right panel). The enriched DNA was quantified by real-time PCR using the SimpleChIP® Universal qPCR Master Mix #88989 and SimpleChIP® Human RPL30 Exon 3 Primers #7014, SimpleChIP® Human GAPDH Exon 1 Primers, and SimpleChIP® Human MyoD1 Exon 1 Primers #4490. The amount of immunoprecipitated DNA in each sample is represented as the signal relative to the total amount of input chromatin, which is equivalent to 1.
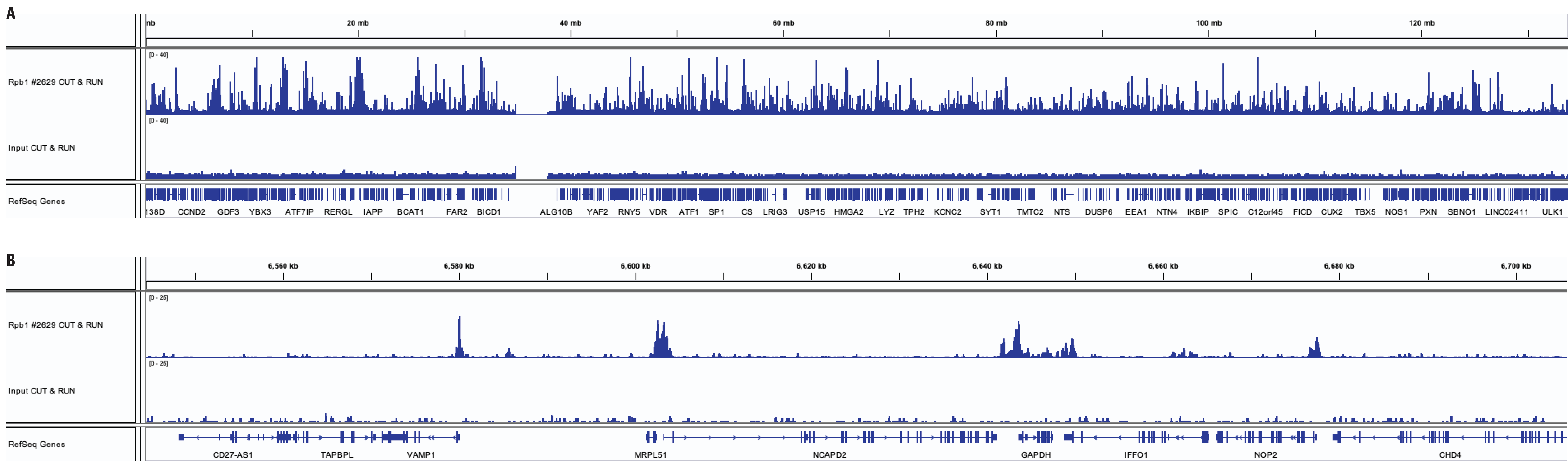
CUT&RUN and ChIP assays were performed with HeLa cells (1x105 cells for CUT&RUN, 4x106 cells for ChIP) and Rpb1 CTD (4H8) Mouse mAb #2629 using the CUT&RUN Assay Kit #86652. DNA libraries were prepared using DNA Library Prep Kit for Illumina (ChIP-seq, CUT&RUN) #56795. Panel A compares enrichment of Rpb1 across chromosome 12, while Panel B compares enrichment at the GAPDH gene, a known target of Rbp1. The input tracks are from the CUT&RUN input sample.
Antibody Versatility
pAG-MNase binds to both mouse and rabbit antibodies, increasing the catalog of antibodies compatible with CUT&RUN assays.
Rabbit Antibody
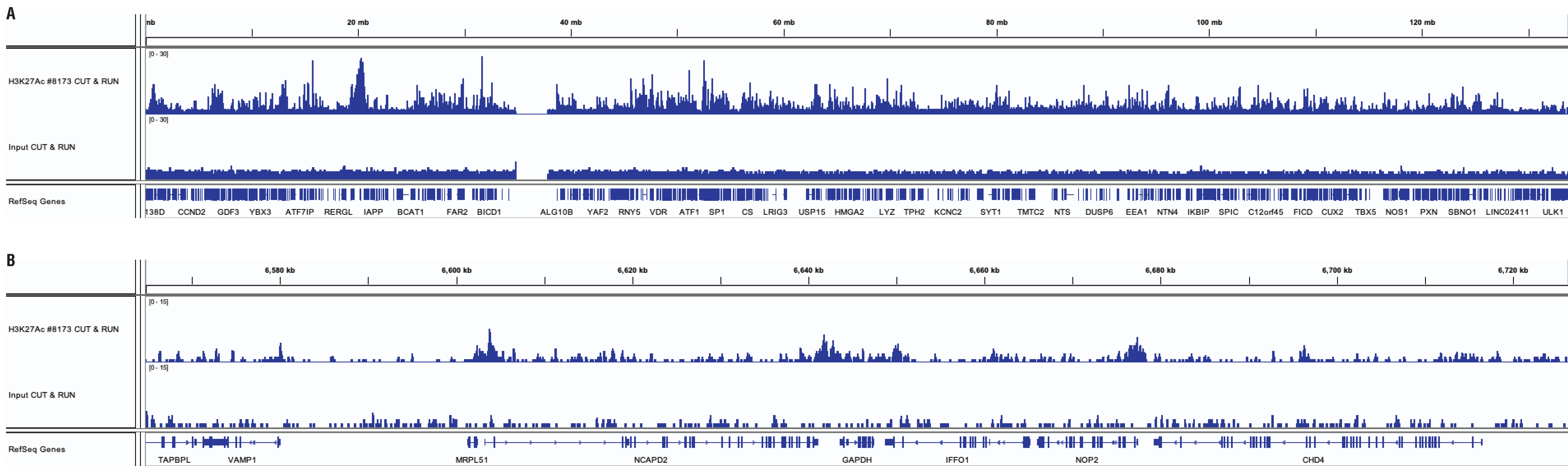
CUT&RUN and ChIP assays were performed with HeLa cells and Acetyl-Histone H3 (Lys27) (D5E4) XP® Rabbit mAb #8173 using the CUT&RUN Assay Kit #86652. DNA libraries were prepared using DNA Library Prep Kit for Illumina (ChIP-seq, CUT&RUN) #56795. Panel A compares enrichment of H3K27Ac across chromosome 12, while Panel B compares enrichment at the GAPDH gene, a known target of H3K27Ac.
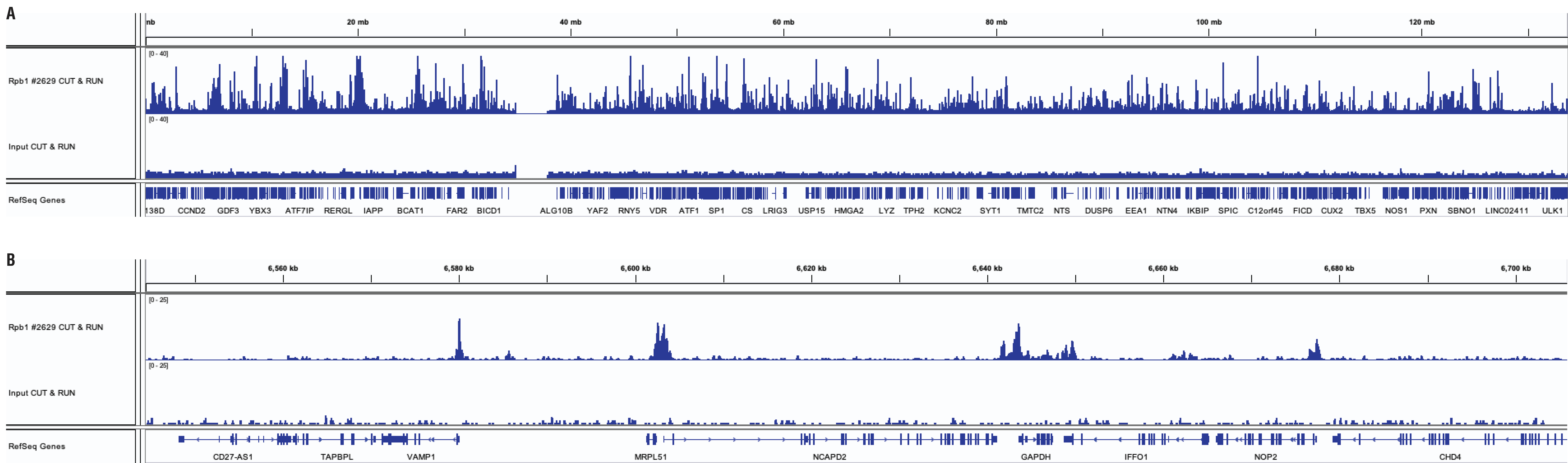
Mouse Antibody
CUT&RUN and ChIP assays were performed with HeLa cells and Rpb1 CTD (4H8) Mouse mAb #2629 using the CUT&RUN Assay Kit #86652. DNA libraries were prepared using DNA Library Prep Kit for Illumina (ChIP-seq, CUT&RUN) #56795. Panel A compares enrichment of Rpb1 across chromosome 12, while Panel B compares enrichment at the GAPDH gene, a known target of Rbp1.
Reproducible Results
Spike in heterologous DNA to normalize your data to improve your experimental reproducibility. Normalize between samples within a given experiment or different experiments.
Normalization of CUT&RUN assay signals using spike-in DNA for qPCR analysis. CUT&RUN assays were performed using the CUT&RUN Assay Kit #86652 with a decreasing number of HCT 116 cells and either Tri-Methyl-Histone H3 (Lys4) (C42D8) Rabbit mAb #9751 (upper panels) or Phospho-Rpb1 CTD (Ser2) (E1Z3G) Rabbit mAb #13499 (lower panels). Enriched DNA was quantified by real-time PCR using the SimpleChIP® Universal qPCR Master Mix #88989 and SimpleChIP® Human GAPDH Exon 1 Primers, SimpleChIP® Human β-Actin Promoter Primers #13653, SimpleChIP® Human β-Actin 3' UTR Primers #13669, and SimpleChIP® Human MyoD1 Exon 1 Primers #4490. The amount of immunoprecipitated DNA in each sample is represented as the signal relative to the total amount of input chromatin (percent input for 100,000 cells). Non-normalized enrichments are depicted in the left panels. The sample normalization spike-in DNA was added into each reaction proportionally to the starting cell number. Based on the difference of qPCR signals from spike-in DNA in each sample, CUT&RUN signals were normalized to the sample containing 100,000 cells. Normalized enrichments are depicted in the right panels.
Avoid “Crosslinking Artifacts”
CUT&RUN works with native chromatin and doesn’t require crosslinking or immunoprecipitation, which can lead to “crosslinking artifacts.”
Product Offerings
CST offers flexible solutions for your CUT&RUN assays, whether you’re just starting to explore epigenetics in your research or you are an expert. All you need to provide is a primary antibody against your protein of interest. CST provides everything else in a simple-to-order CUT&RUN kit that contains all the buffers and reagents you need, along with a detailed protocol. Or just order the pAG-MNase and spike-in DNA if you prefer.
Like all CST products, our CUT&RUN assay products are rigorously validated in-house to ensure that they will work in your experiments and generate data you trust.
Learn how we have validated and optimized our CUT&RUN Kit to yield better signal-to-noise ratios, require fewer cells to analyze protein-DNA interactions, and be compatible with tissue samples. Get up to speed on all the protocol improvements we have made to our CUT&RUN Assay Kit.
Catalog # | Product |
|---|---|
| 86652 | CUT&RUN Assay Kit |
| 40366 | CUT&RUN pAG-MNase and Spike-in DNA |
| 14209 | DNA Purification Buffers and Spin Columns (ChIP, CUT&RUN) |
| 88989 | SimpleChIP® Universal qPCR Master Mix |
| 56795 | DNA Library Prep Kit for Illumina (ChIP-seq, CUT&RUN) #56795 |
| 47538 | Multiplex Oligos for Illumina (Dual Index Primers) (ChIP-seq, CUT&RUN) |
| 29580 | Multiplex Oligos for Illumina (Single Index Primers) (ChIP-seq, CUT&RUN) |
| 66362 | Rabbit (DA1E) mAb IgG XP® Isotype Control (CUT&RUN) |
| 93569 | Concanavalin A Magnetic Beads and Activation Buffer |
| 16359 | Digitonin Solution |
| 27287 | 100X Spermidine |
| 7012 | Protease Inhibitor Cocktail (200X) |
| 7013 | RNAse A (10 mg/ml) |
| 10012 | Proteinase K (20mg/ml) |
| 31415 | CUT&RUN 10X Wash Buffer |
| 15338 | CUT&RUN Antibody Binding Buffer |
| 48105 | CUT&RUN 4X Stop Buffer |
| 42015 | CUT&RUN DNA Extraction Buffer |
| 20533 | 10% SDS Solution |
| 7005 | Glycine Solution (10X) |
| 12606 | 16% Formaldehyde Methanol Free |
| 9872 | PBS-1X pH 7.2 (Sterile) |
References
- Skene PJ, et al. Targeted in situ genome-wide profiling with high efficiency for low cell numbers. (2018) Nat. Protoc 13(5), 1006-1019. Pubmed 29651053
- Meers MP, et al. Improved CUT&RUN chromatin profiling and analysis tools. (2019) BioRxiv 1, 569129. bioRxiv 569129
- Skene PJ, and Henikoff S. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. (2017) Elife 6, e21865. Pubmed 28079019
- Janssens DH, et al. Automated in situ chromatin profiling efficiently resolves cell types and gene regulatory programs. (2018) Epigenetics Chromatin 22(1), 74. Pubmed 30577869