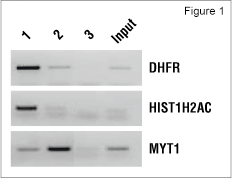
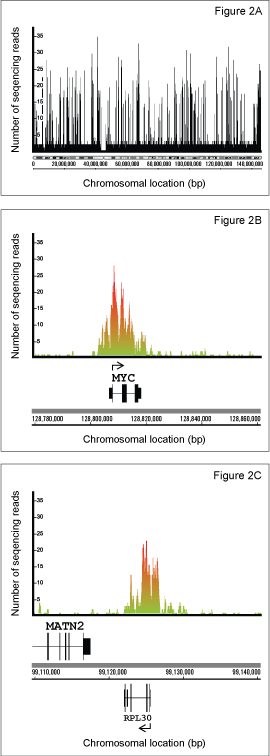
ChIP-Sequencing performed using the SimpleChIP® Kit
Advantages of SimpleChIP® Enzymatic Chromatin IP Kit (Magnetic Beads) for ChIP-Sequencing:
- Enzymatic digestion of chromatin is milder than sonication and better preserves the integrity of the chromatin and antibody epitopes, which means increased IP efficiency.
- Increased IP efficiency means enhanced detection of protein-bound DNA loci.
- ChIP-Grade Protein G Magnetic beads do NOT contain a DNA blocking agent such as salmon sperm DNA, which means no contamination of downstream sequencing.
- ChIP-Grade Protein G Magnetic beads do NOT require centrifugation, which translates to easier sample manipulation and more complete washes.
- ChIP-Grade Protein G Magnetic beads provide greater sensitivity with lower background, which is essential when detecting DNA bound transcription factors and cofactors.
SimpleChIP® Kit used with ChIP-Sequencing
Epigenetic landscapes consist of distinct domains of euchromatin and heterochromatin, which are unique to different cell types and different stages in development. Epigenetic regulation of gene expression is mediated through posttranslational modifications of histones. For example, tri-methylation of histone H3 Lys4 is a modification associated with active chromatin (euchromatin), while tri-methylation of histone H3 Lys27 is a mark of inactive heterochromatin.
A powerful method used to identify localized regions of histone modifications as well as binding sites for transcription factors on a genome-wide scale is the chromatin immunoprecipitation (ChIP) assay. The ChIP assay uses formaldehyde to covalently crosslink proteins to their DNA substrates in living cells. The protein:DNA complexes are then immunoprecipitated using antibodies generated against specific histone modifications or transcription factors. The precipitated DNA is purified and can be analyzed by hybridization to oligo-nucleotide microarrays (ChIP-chip) or by high throughput DNA sequencing (ChIP-seq). These methods provide an opportunity to take a snapshot of DNA-protein interactions in a given cell type, using populations of cultured cells, subsets of cells taken at specific times of the cell cycle or development, or even cells taken directly from tissue samples. ChIP-chip was the first method used for whole genome binding site analyses. However, multiple DNA microarrays are required to cover the entire human genome, resulting in high costs for comprehensive studies. ChIP-seq technology is another method that offers the ability to identify binding sites across the entire genome in a single sequencing run. This can be done using the Illumina GA2 sequencer, which generates short sequence reads that are sufficient for accurate mapping of the enriched DNA fragments to their genomic location.
Using the SimpleChIP® Enzymatic Chromatin IP Kit (Magnetic Beads) #9003 from Cell Signaling Technology, we have performed ChIP-seq experiments to identify the epigenetic signatures of tri-methyl-histone H3 Lys4 and tri-methyl-histone H3 Lys27 histone marks in the K562 erythroleukemia cell line. Chromatin was prepared from K562 cells as described in the SimpleChIP® protocol and immunoprecipitated with Tri-Methyl-Histone H3 (Lys4) (C42D8) Rabbit mAb #9751, Tri-Methyl-Histone H3 (Lys27) (C36B11) Rabbit mAb #9733 and Normal Rabbit IgG #2729 as a negative control. The enrichment of tri-methyl histone H3 Lys4 and tri-methyl histone H3 Lys27 at known binding sites was confirmed by standard polymerase chain reaction (Figure 1). The immunoprecipitated DNA was then used to prepare libraries for sequencing with the Illumina GA2 sequencing platform. Obtained sequences were mapped to UCSC Human Genome Assembly (HG18) and only uniquely mapped sequences were retained. Enriched binding sites are reflected by elevated number of sequence reads giving rise to peaks (Figure 2). The rabbit monoclonal antibodies were found to give excellent enrichment, as reflected in the average peak height of 17 sequence reads and very low background.
CST would like to thank H. O'geen and P.J. Farnham of University of California, Davis in Davis, California for sharing their ChIP-Sequencing data.