CUT&Tag Frequently Asked Questions
Questions
- Is CUT&Tag assay compatible with spike-in sample normalization?
- Is there a maximum number of cells that can be used in a single CUT&Tag assay?
- What is the fewest number of cells I can use in a reaction with the CST CUT&Tag kit?
- Do you observe any differences in assay performance among different cell lines?
- Do you have any specific advice for working with adherent cell lines? How about suspension cell lines?
- Is it possible to perform CUT&Tag on fixed cells?
- Is it possible to perform CUT&Tag on frozen cells?
- Is it possible to perform CUT&Tag on tissue samples like heart, brain, or developing mouse tissues?
- Is it possible to perform CUT&Tag on frozen tissue samples?
- Should I be concerned that my Concanavalin A beads "clump together" in some of my CUT&Tag experiments?
- Do I need to optimize the Tn5 tagmentation conditions for different cell types?
- Should I include digitonin in my wash buffers for the entire experiment? Will it be too stressful for the cells?
- Does the CST CUT&Tag kit work with isolated nuclei?
- Does the CUT&Tag assay show a bias toward euchromatin or heterochromatin?
- What controls can I use to show my CUT&Tag experiment is working?
- What is the best way to choose an antibody for my CUT&Tag assay if I can not find a CUT&Tag-validated antibody for my target protein?
- Which method is better to use for purifying CUT&Tag DNA, a phenol-chloroform DNA extraction using ethanol precipitation or DNA affinity columns?
- Can CUT&Tag-enriched DNA be analyzed by qPCR, or do I have to do next-generation sequencing (NGS) to analyze my data?
- What is the expected yield of CUT&Tag DNA libraries?
- I would like to QC and analyze the fragment sizes of my CUT&Tag DNA before sequencing, but my yield is too low to see on an Agilent Bioanalyzer or TapeStation system. What should I do?
- How do I pool CUT&Tag DNA libraries with a variety of yields?
- What DNA library prep kit do you recommend using for the CUT&Tag assay?
- Do you recommend performing size selection of the DNA during the library preparation?
- What if the whole CUT&Tag kit is left at -20°C or room temperature upon receipt?
Is CUT&Tag assay compatible with spike-in sample normalization?
Yes. We recommend the Drosophila spike-in normalization approach, which enables reliable and consistent normalization across samples, corrects for variations in starting cell number, technical variation among samples during processing, and non-specific enrichment coming from isotype control antibodies. Detailed instructions for implementing spike-in controls are provided in our Drosophila Spike-In Control Kit for CUT&Tag (Rabbit) #29811 or Drosophila Spike-In Control Kit for CUT&Tag (Mouse) #19629.
With the Drosophila spike-in strategy, Drosophila nuclei are added concurrently with the test cells at the start of the CUT&Tag workflow, ensuring they experience all the same steps and conditions as the test cells. The H2Av Rabbit or Mouse Monoclonal Antibody, which specifically recognizes Drosophila chromatin, is included along with the test antibody. A normalization factor is then generated from the Drosophila enrichment signal and applied to the test genome to normalize signal across samples. This approach normalizes all steps throughout the entire protocol.
Some protocols rely on contaminating E. coli DNA present in pAG-Tn5 for normalization, but this can lead to inconsistencies when switching enzyme lots, as the amount of contaminating DNA varies. The Drosophila spike-in strategies provide better lot-to-lot consistency and more reliable normalization.
Is there a maximum number of cells that can be used in a single CUT&Tag assay?
We have found 100,000 cells per assay to be sufficient for enriching target loci for multiple histone modifications, transcription factors, and transcription cofactors. However, one can increase the number of cells to 250,000 per assay without scaling up beads, antibodies, enzyme, or buffers.
What is the fewest number of cells I can use in a reaction with the CST CUT&Tag kit?
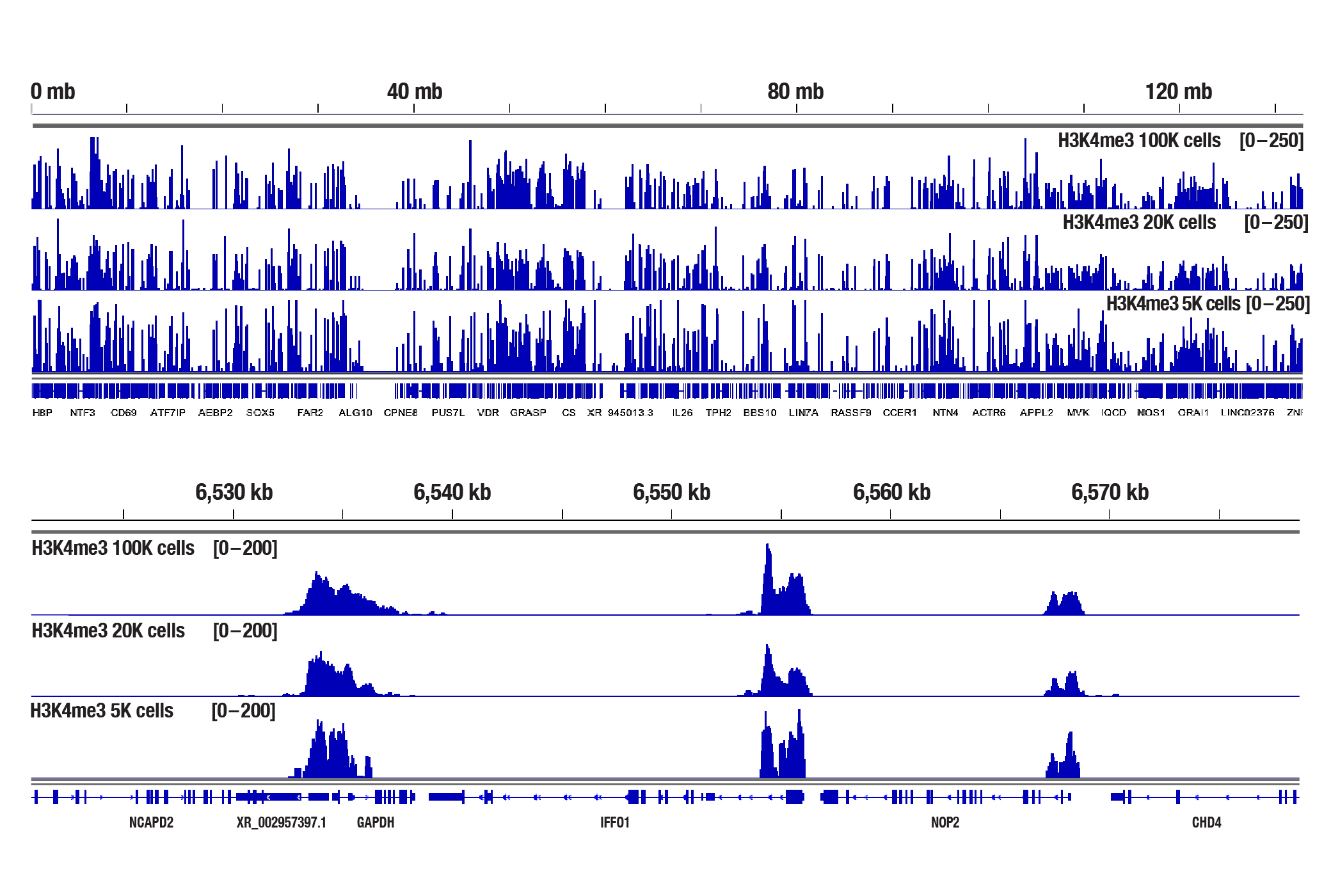
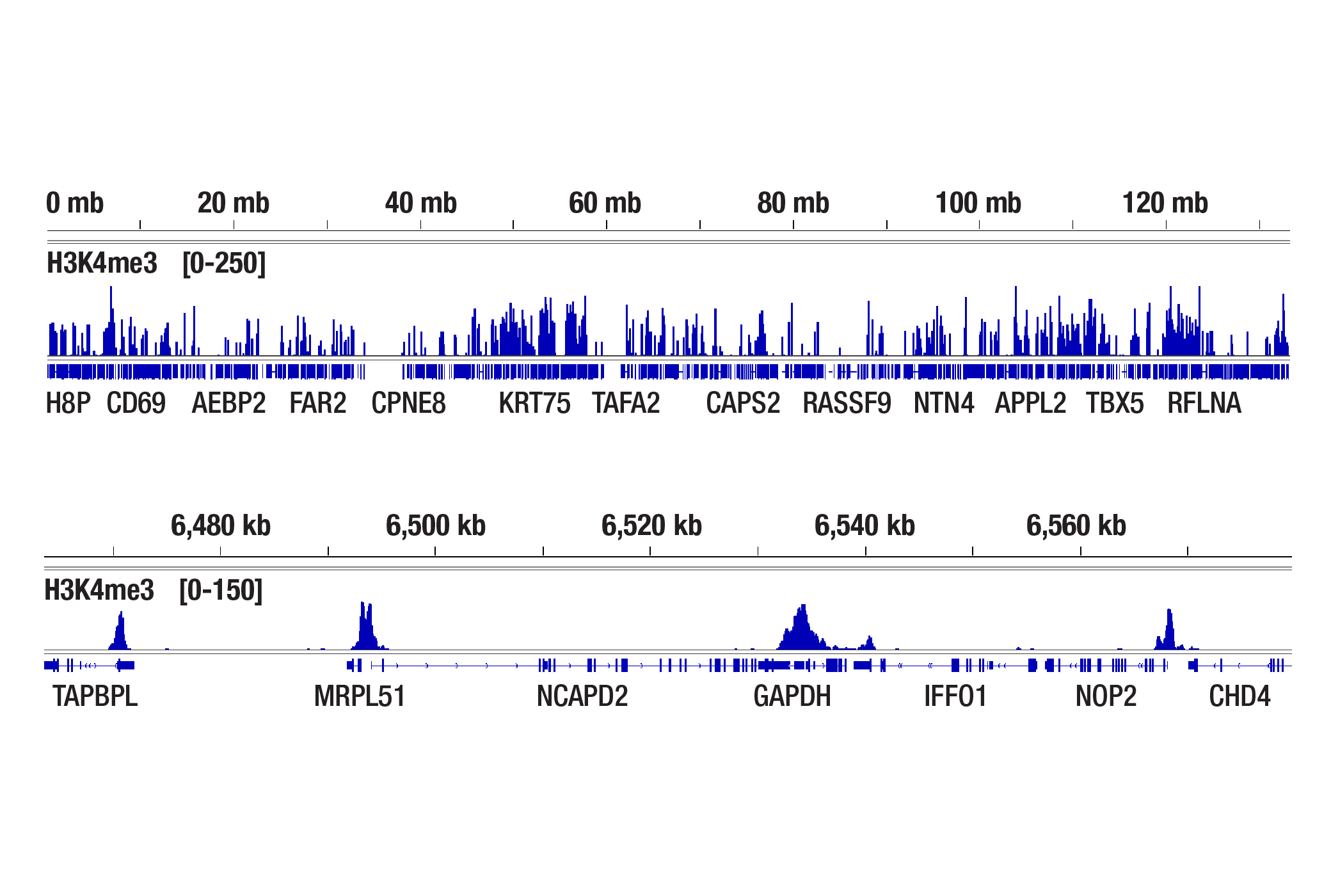
We have shown that our CUT&Tag Assay Kit #77552 works with as few as 5,000-10,000 cells for histone modifications and 20,000 cells for transcription factors and cofactors (see images).
The CUT&Tag assay was performed with 100,000, 20,000, or 5,000 NCCIT cells (as indicated) and Tri-Methyl-Histone H3 (Lys4) (C42D8) Rabbit mAb #9751, using the CUT&Tag Assay Kit #77552. DNA libraries were prepared using the CUT&Tag Dual Index Primers and PCR Master Mix for Illumina Systems #47415. The upper panel compares enrichment of H3K4me3 across chromosome 12, while the lower panel compares enrichment around GAPDH, a known target gene of H3K4me3.
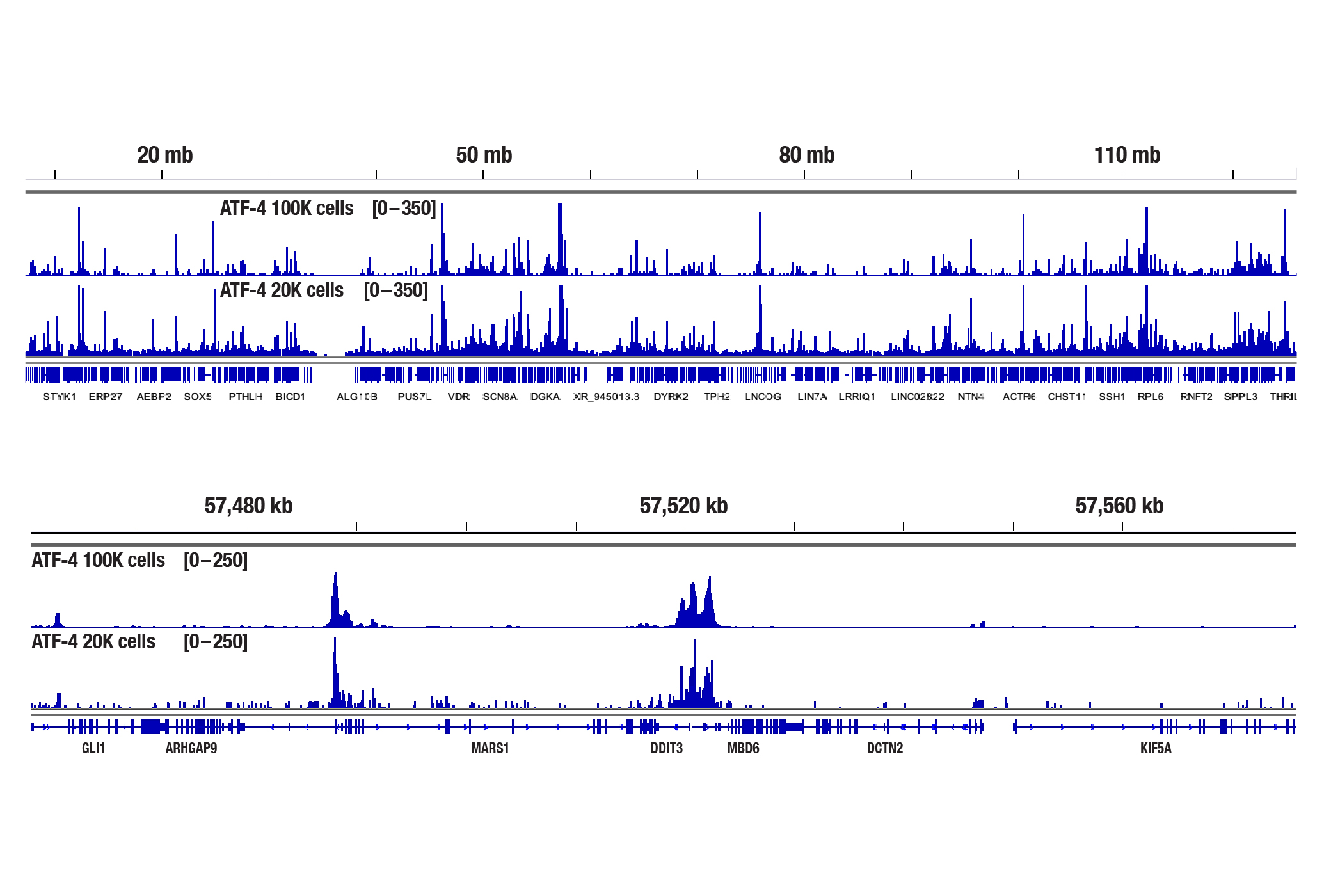
The CUT&Tag assay was performed with 100,000 or 20,000 Hep G2 cells (as indicated) treated with Thapsigargin #12758 (300 nM) for 4 hr and ATF-4 (D4B8) Rabbit mAb #11815, using the CUT&Tag Assay Kit #77552. DNA libraries were prepared using the CUT&Tag Dual Index Primers and PCR Master Mix for Illumina Systems #47415. The upper panel compares enrichment of ATF-4 across chromosome 12, while the lower panel compares enrichment around DDIT3/CHOP, a known target gene of ATF-4.
Do you observe any differences in assay performance among different cell lines?
We have performed CUT&Tag on dozens of cultured adherent and suspension cell lines, both human and mouse. So far we have not noticed any significant difference in performance between these cell lines.
Do you have any specific advice for working with adherent cell lines? How about suspension cell lines?
With adherent cell lines, one first needs to detach the cells from the dish. We recommend using trypsin to detach the cells. Alternatively, you can use accutase, but the resulting cell pellet doesn't pack as well during centrifugation, so you tend to lose cells during the wash steps. Suspension cells are easier to manage. You can count the cells using a hemacytometer or auto-cell counter and then collect the number of cells you need. We don't observe a significant difference in assay performance with adherent versus suspension cells.
Is it possible to perform CUT&Tag on fixed cells?
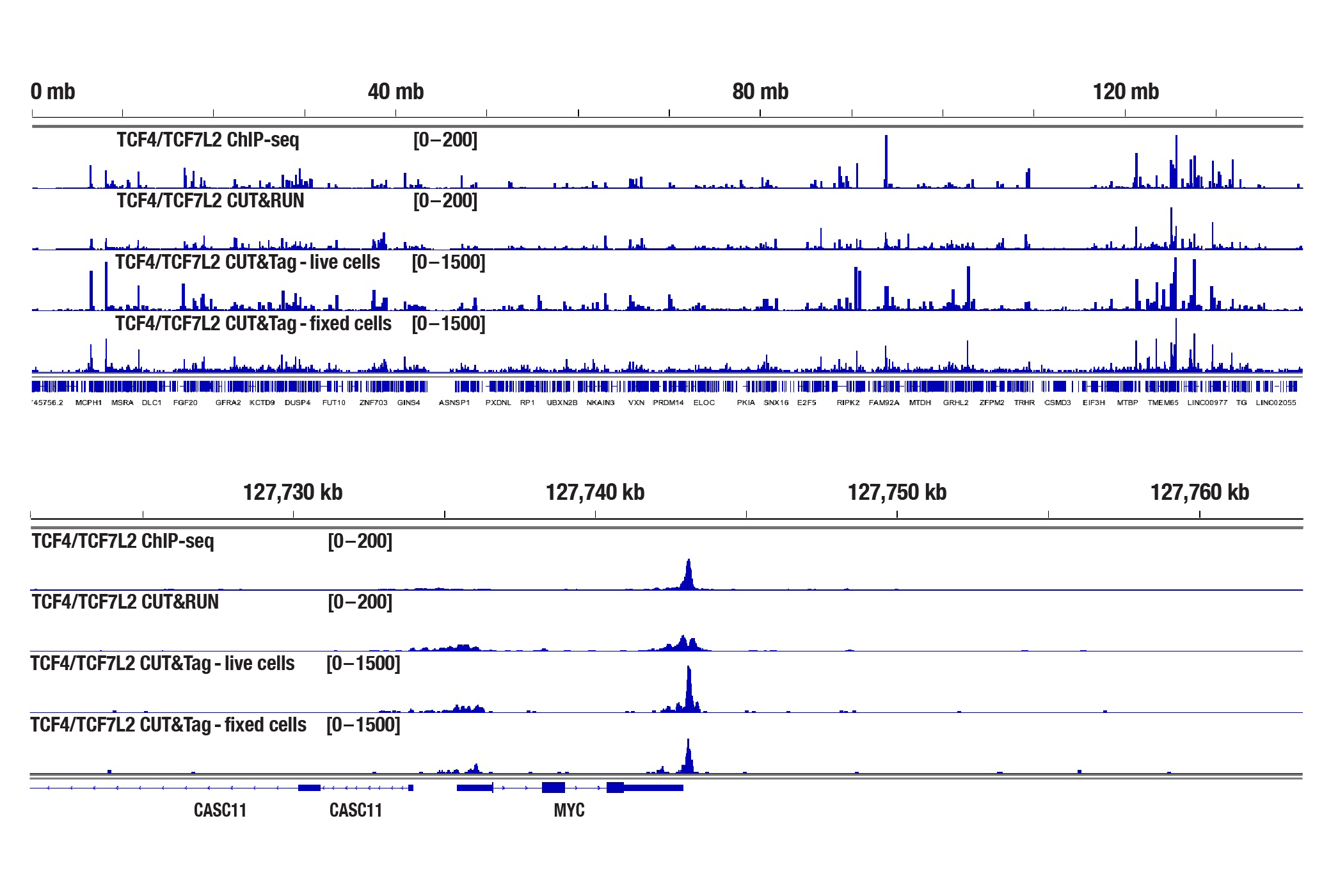
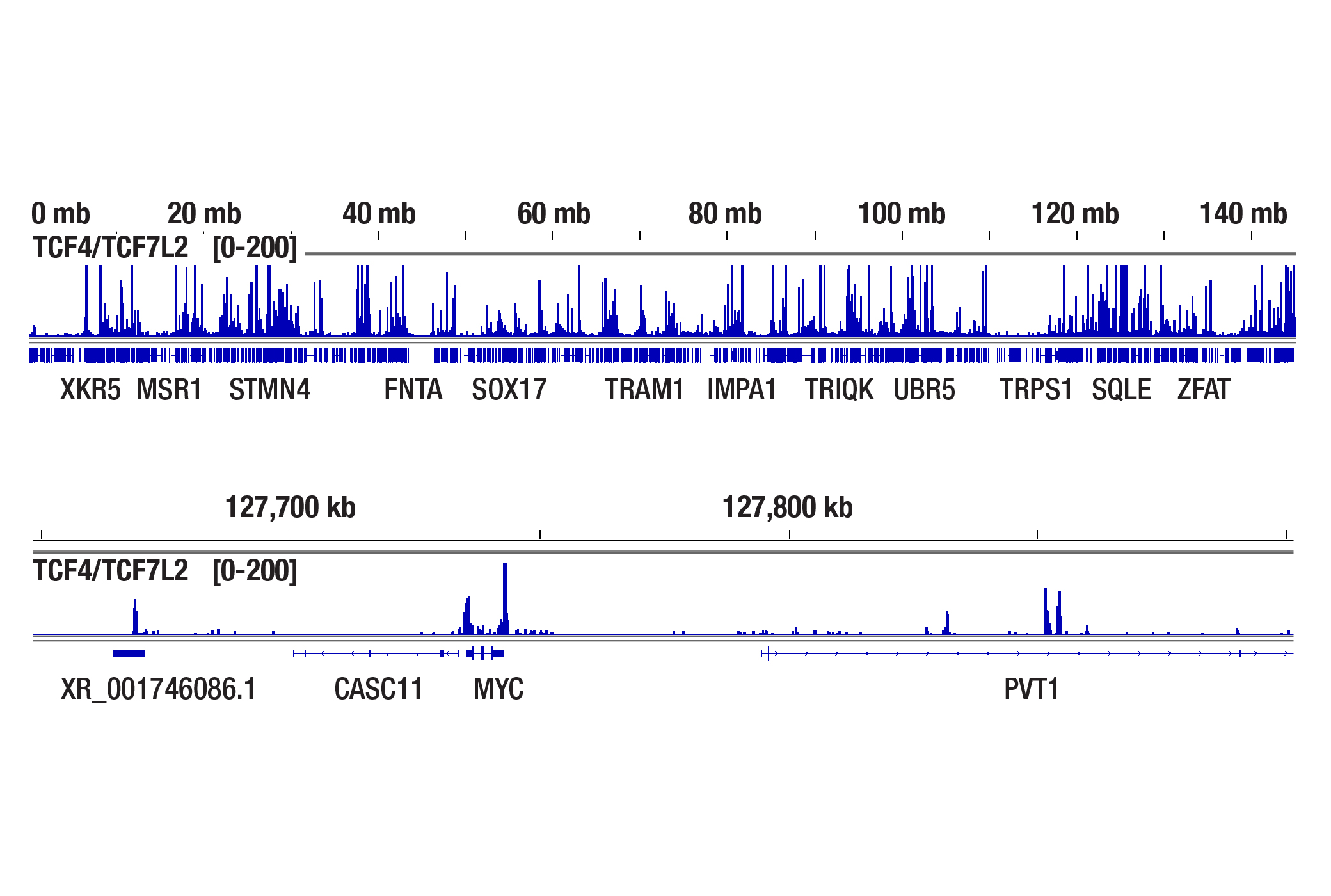
Yes. Cell fixation is recommended when the cells are very fragile (easily lysed during experiments), when the cellular signaling pathway is sensitive to the Concanavalin A beads used for cell binding, or when you hope to freeze the cell pellets for future use. If fixation is necessary, refer to the CUT&Tag Assay Kit Protocol, Appendix A to lightly fix cells prior to the CUT&Tag experiment. We find that live cells produce better results than fixed cells in the CUT&Tag assay (see image). Therefore, we recommend using live cells, if possible, unless fixation is necessary for the scenarios mentioned above.
ChIP-seq, CUT&RUN, and CUT&Tag assays were performed with HCT 116 cells and TCF4/TCF7L2 (C48H11) Rabbit mAb #2569, using the SimpleChIP® Plus Enzymatic Chromatin IP Kit (Magnetic Beads) #9005, CUT&RUN Assay Kit #86652, or CUT&Tag Assay Kit #77552. Cells for the CUT&Tag assay were fixed using 0.1% formaldehyde for 2 min. DNA libraries were prepared using the DNA Library Prep Kit for Illumina Systems (ChIP-seq, CUT&RUN) #56795 for ChIP-seq and CUT&RUN samples and the CUT&Tag Dual Index Primers and PCR Master Mix for Illumina Systems #47415 was used for CUT&Tag samples. The figures show enrichment of TCF4/TCF7L2 across chromosome 8 (upper), including MYC (lower), a known target gene of TCF4/TCF7L2.
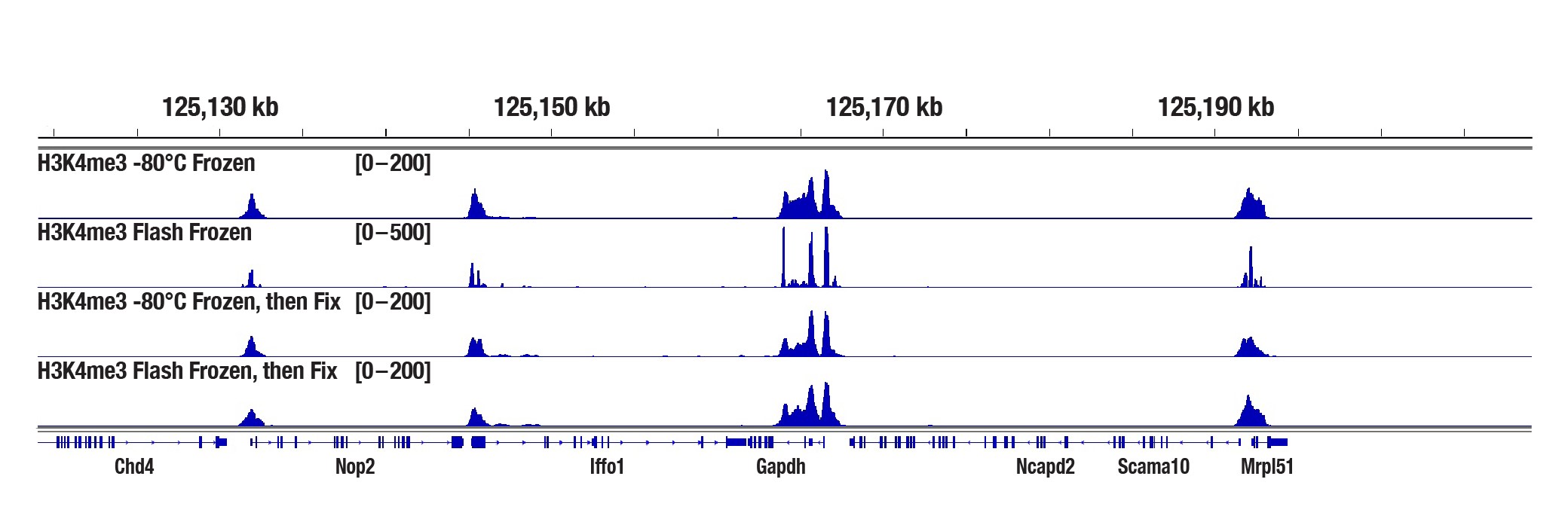
Is it possible to perform CUT&Tag on frozen cells?
Yes. CUT&Tag can be performed on frozen cell pellets, with or without fixation prior to freezing. Frozen or fixed-frozen cell pellets can be stored at -80° C for up to 6 months. In our experience, live cells generally yield better performance than frozen or fixed-frozen cell pellets, so we recommend using live cells whenever possible unless frozen samples are required for certain situations.
Is it possible to perform CUT&Tag on tissue samples like heart, brain, or developing mouse tissues?
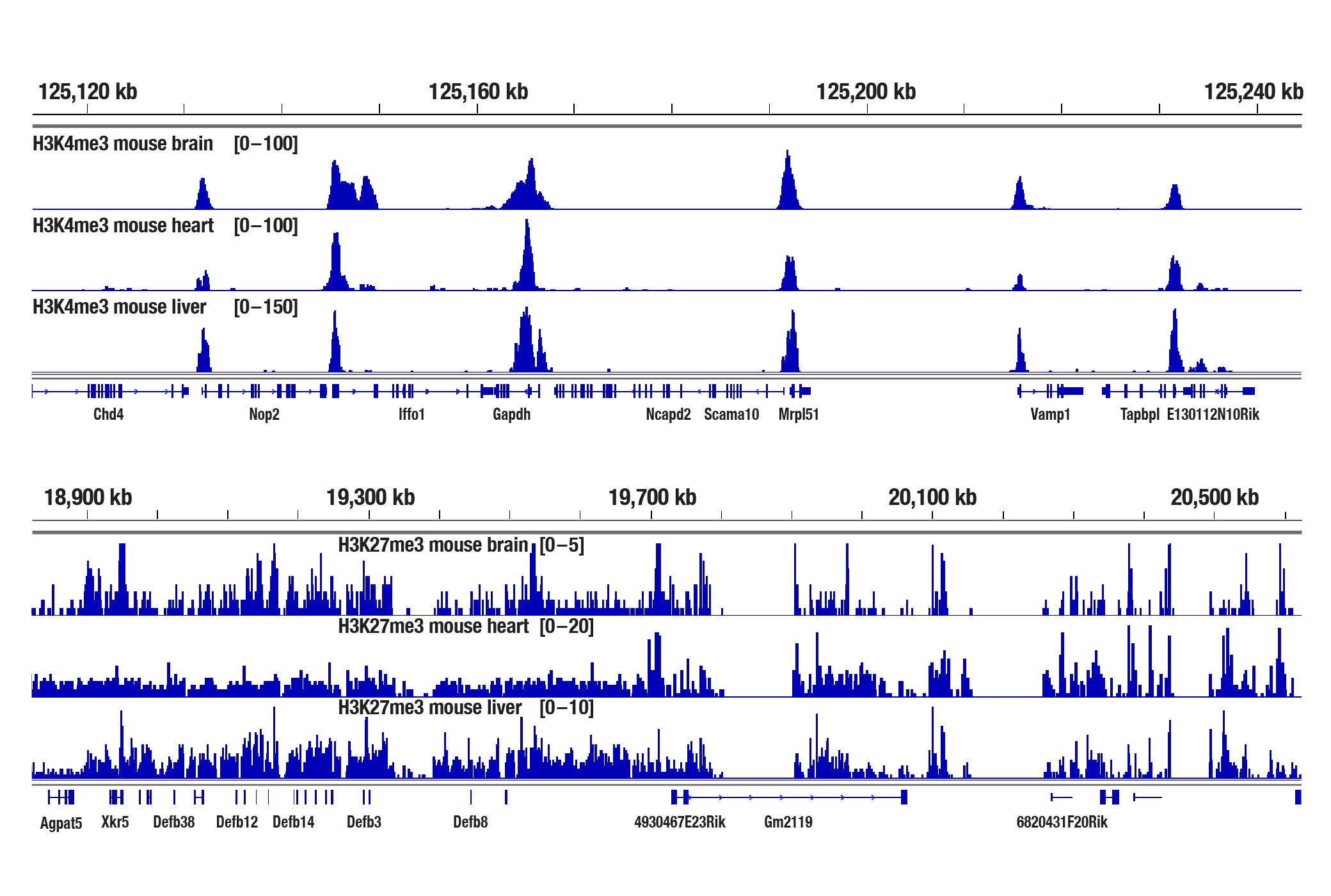
Yes. We have shown that our CUT&Tag Assay Kit #77552 works with mouse liver, mouse brain, and mouse heart tissues for histone targets with as little as 1 mg of tissue for each CUT&Tag reaction (see image). However, non-histone targets such as transcription factors and cofactors are not well enriched in tissues in the CUT&Tag assay. For analysis of transcription factors and cofactors in tissues, we recommend using the CUT&RUN Assay Kit #86652. Fresh tissues typically generate comparable or stronger CUT&Tag signals than fixed tissues. If fixation is necessary, refer to the CUT&Tag Assay Kit Protocol, Appendix B for the light fixation of tissues prior to the CUT&Tag experiment. Fixed tissues can be frozen for up to 6 months before use in the CUT&Tag assay.
The CUT&Tag assay was performed with 1 mg of fresh mouse brain, heart, or liver tissues (as indicated) and either Tri-Methyl-Histone H3 (Lys4) (C42D8) Rabbit mAb #9751 or Tri-Methyl-Histone H3 (Lys27) (C36B11) Rabbit mAb #9733, using the CUT&Tag Assay Kit #77552. DNA libraries were prepared using the CUT&Tag Dual Index Primers and PCR Master Mix for Illumina Systems #47415. The upper panel shows enrichment of H3K4me3 around its known target gene Gapdh, while the lower panel shows enrichment of H3K27me3 around Defb genes.
Is it possible to perform CUT&Tag on frozen tissue samples?
Yes, our CUT&Tag Assay Kit works with flash frozen tissue samples or tissue samples directly frozen at -80°C. After thawing, fixation of the tissue sample is optional, as we haven't found that fixation boosts the CUT&Tag signal strength (see image).
Mouse liver tissue was either frozen at -80°C or frozen in liquid nitrogen for 2 min before storing at -80°C (flash frozen), with or without a 10 min fixation in 0.1% formaldehyde after thawing, as indicated. The CUT&Tag assay was performed with 2.5 mg of tissue and Tri-Methyl-Histone H3 (Lys4) (C42D8) Rabbit mAb #9751, using the CUT&Tag Assay Kit #77552. DNA libraries were prepared using the CUT&Tag Dual Index Primers and PCR Master Mix for Illumina Systems #47415. The panel shows binding across Gapdh, a known target gene of H3K4me3.
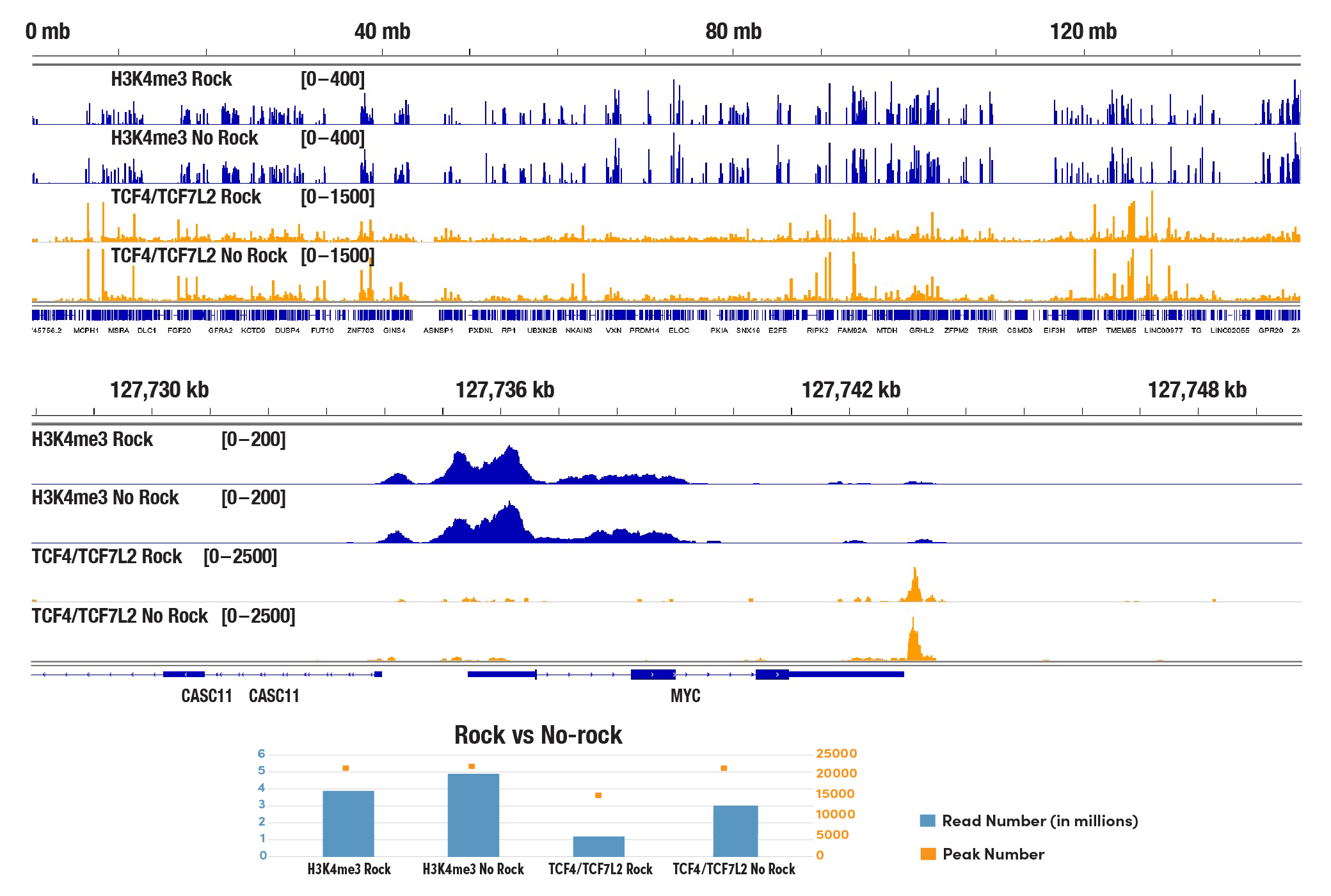
Should I be concerned that my Concanavalin A beads "clump together" in some of my CUT&Tag experiments?
The Concanavalin A beads can clump together rather easily, especially when the cells become lysed. Make sure your cells are healthy and be sure to treat them very gently during the cell wash. We also recommend limiting the room temperature incubation time of the cell suspension with the Concanavalin A beads to 5 minutes. Resting the tubes at the appropriate incubation temperatures in lieu of rotating or rocking the tubes during the incubations for cell-bead binding, antibody binding, and pAG-Tn5 enzyme binding helps to mitigate the beads from clumping and does not have any adverse effects on sequencing depth, identified peak numbers, or the binding patterns (see image). With all that being said, our internal testing has shown that bead clumping does not negatively affect the final results of CUT&Tag experiments.
The CUT&Tag assay was performed with 100,000 HCT 116 cells and either Tri-Methyl-Histone H3 (Lys4) (C42D8) Rabbit mAb #9751 or TCF4/TCF7L2 (C48H11) Rabbit mAb #2569, using the CUT&Tag Assay Kit #77552. Sample tubes were either rocked during cell:Concanavalin A bead binding, antibody incubations, and tagmentation (indicated as Rock) or stood still in a tube rack at appropriate temperatures for all mentioned steps (indicated as No Rock). DNA libraries were prepared using the CUT&Tag Dual Index Primers and PCR Master Mix for Illumina Systems #47415. All four samples are pooled and sequenced together. The lower panel shows the reads number obtained from each indicated sample and the peak number identified in each dataset. The upper panel shows enrichment of H3K4me3 and TCF4/TCF7L2 across chromosome 8 (upper), including MYC (lower), a known target gene of H3K4me3 and TCF4/TCF7L2.
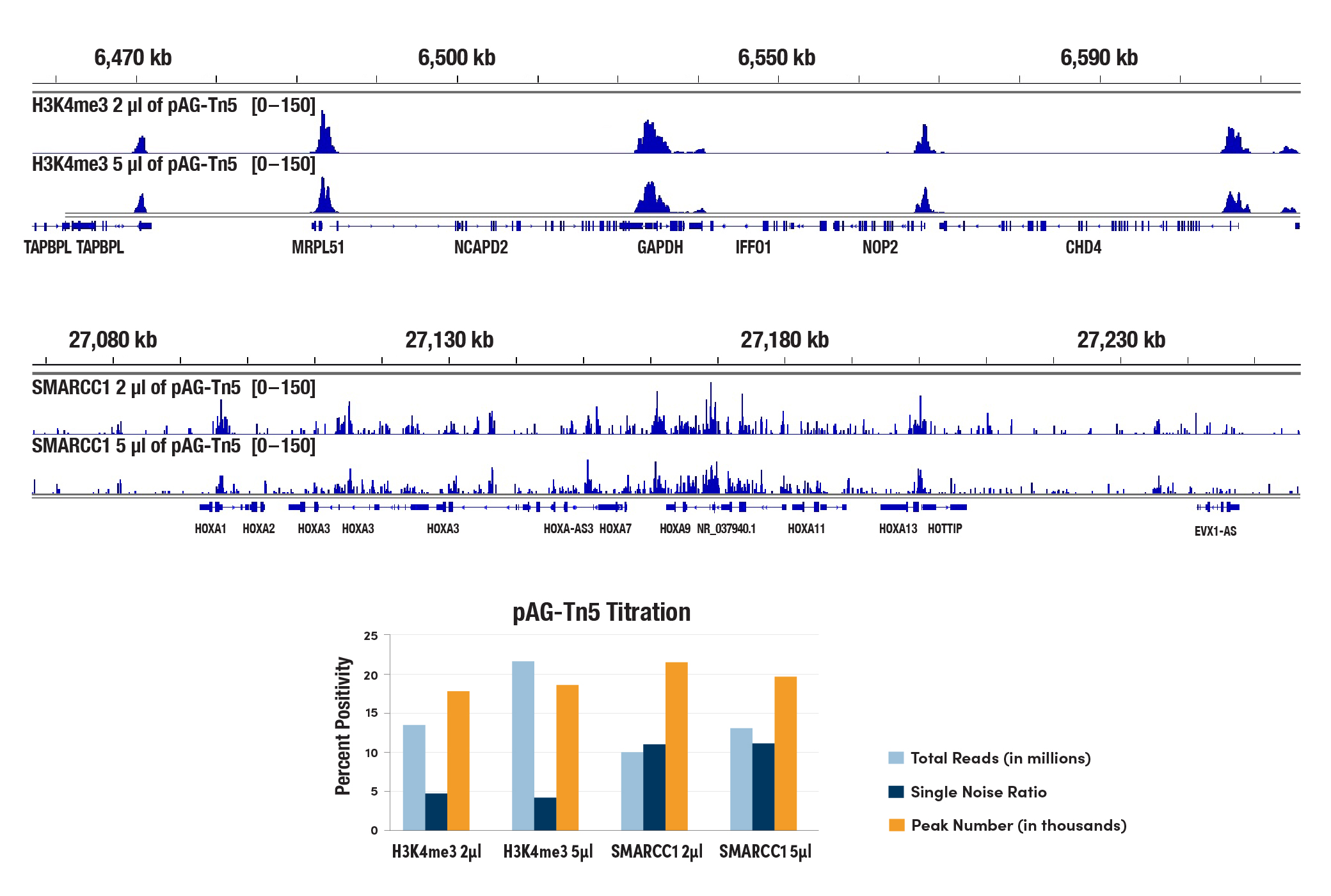
Do I need to optimize the Tn5 tagmentation conditions for different cell types?
Our optimized tagmentation conditions for the CUT&Tag assay work well across multiple cell lines, both adherent and suspension. We recommend using 2 μL of CUT&Tag pAG-Tn5 (Loaded) #79561 in each 50 μL of tagmentation reaction. Using a larger volume of pAG-Tn5 in each reaction doesn't increase the number of reads, peak number, or signal-to-noise ratio in our in-house tests (see image). In fact, pAG-Tn5 activity is more critical to the success of the CUT&Tag assay than amount of pAG-Tn5 used. Please make sure to store the pAG-Tn5 enzyme properly at -20°C and do not use expired pAG-Tn5 as activity does decline over time. The shelf life of CUT&Tag pAG-Tn5 (Loaded) #79561 is 6 months. Additionally, we have optimized the salt concentration in our tagmentation buffer to ensure efficient target-specific tagmentation and to limit non-specific tagmentation at open chromatin regions.
The CUT&Tag assay was performed with 100,000 HCT 116 cells with either 2 µL or 5 µL of pAG-Tn5 (Loaded) #79561 used in each reaction, and either Tri-Methyl-Histone H3 (Lys4) (C42D8) Rabbit mAb #9751 or SMARCC1/BAF155 (D7F8S) Rabbit mAb #11956 (as indicated), using the CUT&Tag Assay Kit #77552. DNA libraries were prepared using the CUT&Tag Dual Index Primers and PCR Master Mix for Illumina Systems #47415. All four samples were pooled and sequenced together. The lower panel shows that the number of reads, the signal-to-noise ratio across the genome, and the peak number identified from each indicated sample are comparable between the two different volumes of pAG-Tn5. The upper panel shows H3K4me3 binding across GAPDH (upper), a known target gene of H3K4me3, and SMARCC1 binding across HoxA gene clusters (lower), known target genes of SMARCC1.
Should I include digitonin in my wash buffers for the entire experiment? Will it be too stressful for the cells?
Yes, the digitonin needs to be present in all of the buffers during the entire experiment because the membrane permeabilization caused by digitonin is reversible. Removing or lowering the amount of digitonin in the buffers may interfere with both the antibodies and pAG-Tn5 entering the nuclei. Cells are more sensitive to digitonin concentration rather than the digitonin treatment time. If you find that the recommended amount of digitonin is leading to extensive lysis of your cells, you can optimize the concentration of digitonin in your assay as described in the CUT&Tag Assay Kit Protocol, Appendix C.
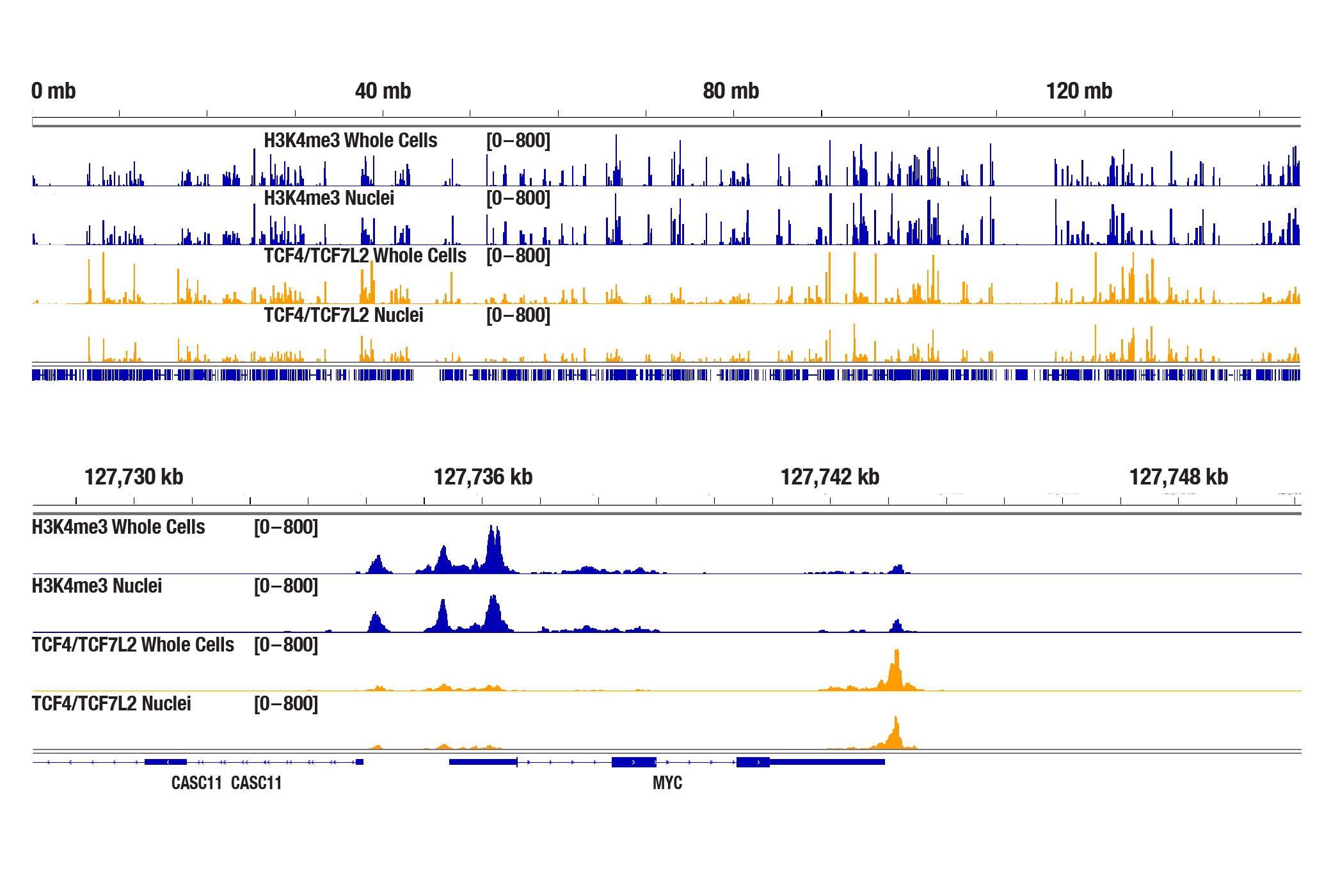
Does the CST CUT&Tag kit work with isolated nuclei?
Our CUT&Tag kit is optimized for use with whole cells and works with a large number of cell lines, both adherent and suspension. While our kit works with pre-isolated nuclei because the Concanavalin A beads bind to glycoproteins on the surface of both cells and nuclei, we haven't found that pre-isolation of cell nuclei helps to increase the CUT&Tag signal (see image). Pre-isolation of cell nuclei should only be necessary for cells that are not efficiently permeabilized by digitonin, such as yeast or plant cells, which both contain cell walls.
The CUT&Tag assay was performed with 100,000 whole cells or pre-isolated nuclei (as indicated) from HCT 116 cells and either Tri-Methyl-Histone H3 (Lys4) (C42D8) Rabbit mAb #9751 or TCF4/TCF7L2 (C48H11) Rabbit mAb #2569, using the CUT&Tag Assay Kit #77552. DNA libraries were prepared using the CUT&Tag Dual Index Primers and PCR Master Mix for Illumina Systems #47415. The figures show enrichment of H3K4me3 and TCF4/TCF7L2 across chromosome 8 (upper), including MYC (lower), a known target gene of H3K4me3 and TCF4/TCF7L2.
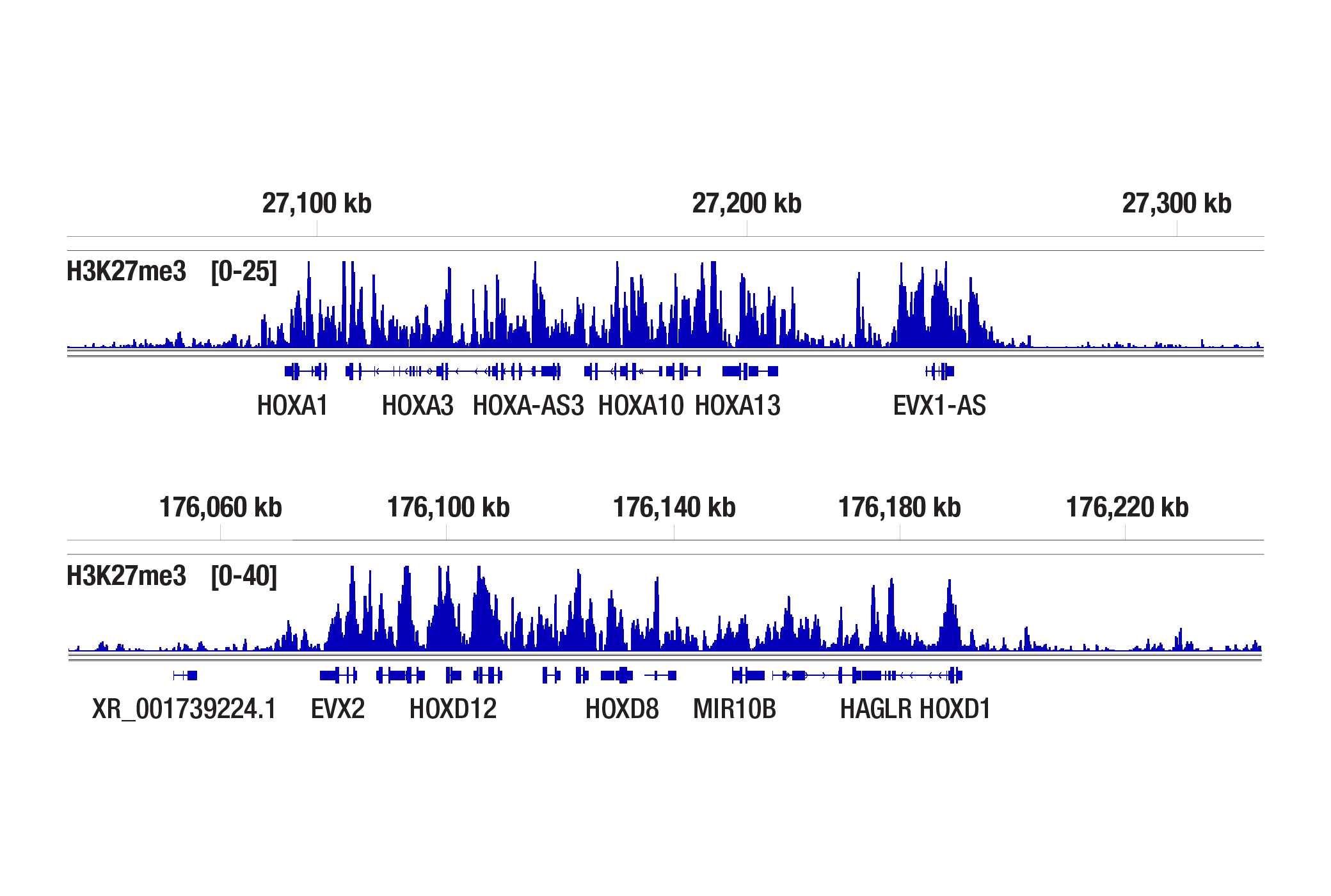
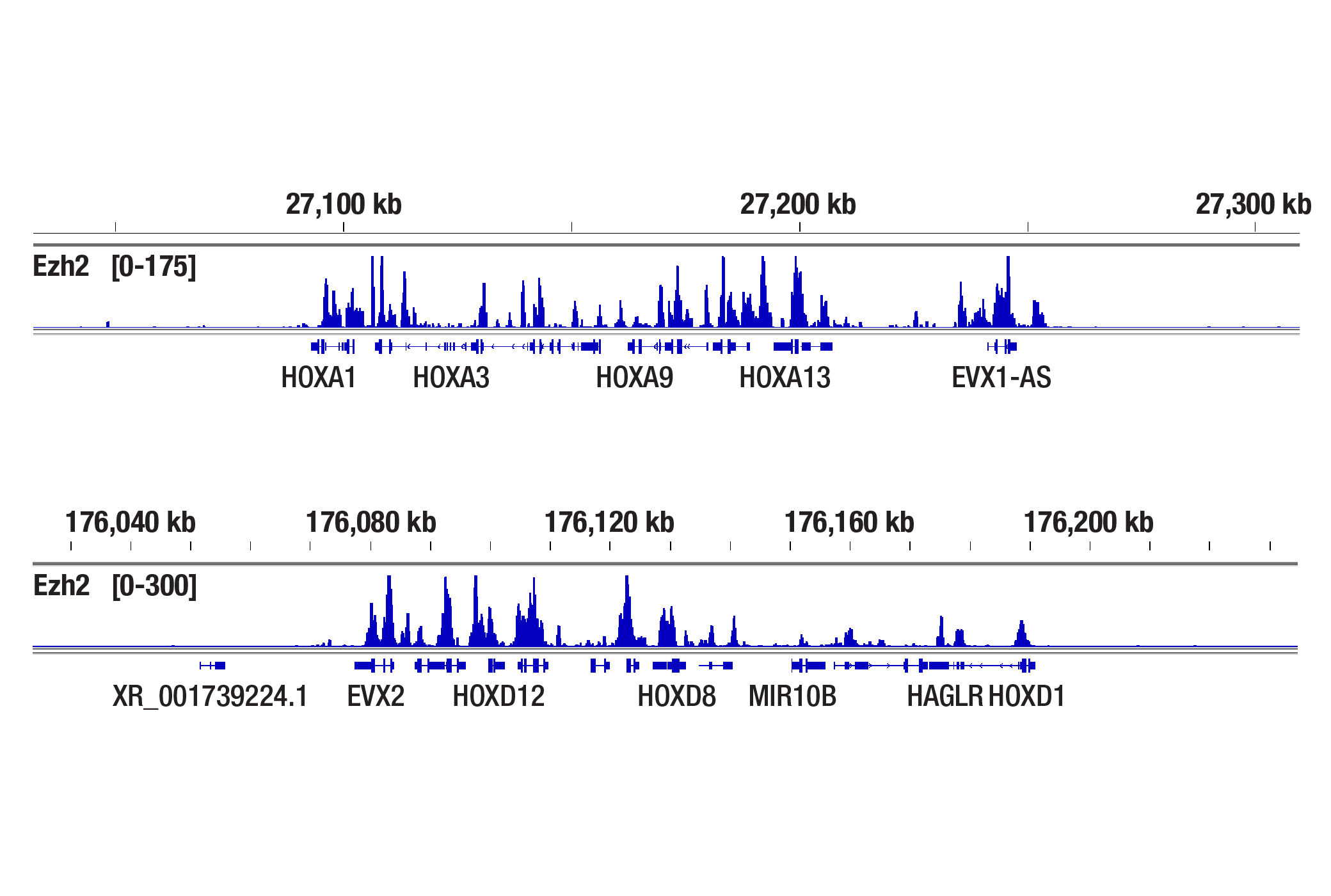
Does the CUT&Tag assay show a bias toward euchromatin or heterochromatin?
With the appropriate salt concentration in the tagmention buffer, we don't observe bias toward euchromatin or heterochromatin in our assays. In CUT&Tag, the target-specific antibody and the secondary antibody recruit the pAG-Tn5 and direct the tagmentation to the chromatin directly adjacent to the target-specific antibody, whether in euchromatin or heterochromatin. The active tethering of pAG-Tn5 to the chromatin allows for tagmentation to occur even in less accessible heterochromatin regions. If CST scientists observe non-specific tagmentation at open chromatin regions when they are validating an antibody for CUT&Tag, then the antibody is not approved for use in the CUT&Tag assay. Our CUT&Tag Assay Kit #77552 has been shown to work well with antibodies against active, accessible euchromatic histone modifications (see Tri-Methyl-Histone H3 (Lys4) (C42D8) Rabbit mAb #9751) and activating transcription factors (see TCF4/TCF7L2 (C48H11) Rabbit mAb #2569), as well as repressive, inaccessible heterochromatic histone modifications (see Tri-Methyl-Histone H3 (Lys27) (C36B11) #9733) and polycomb repressor complex proteins that are associated with inactive heterochromatin (see Ezh2 (D2C9) XP® Rabbit mAb #5246) (see images).
The CUT&Tag assay was performed with HCT 116 cells and Tri-Methyl-Histone-H3 (Lys4) (C42D8) Rabbit mAb #9751 using the CUT&Tag Assay Kit #77552. DNA libraries were prepared using the CUT&Tag Dual Index Primers and PCR Master Mix for Illumina Systems #47415. The figures show binding across chromosome 12 (upper), including GAPDH (lower), a known target gene of H3K4me3.
The CUT&Tag assay was performed with HCT 116 cells and TCF4/TCF7/L2 (C48H11) Rabbit mAb #2569 using the CUT&Tag Assay Kit #77552. DNA libraries were prepared using the CUT&Tag Dual Index Primers and PCR Master Mix for Illumina Systems #47415. The figures show binding across chromosome 8 (upper), including MYC (lower), a known target gene of TCF4/TCF7L2.
The CUT&Tag assay was performed with NCCIT cells and Tri-Methyl-Histone H3 (Lys27) (C36B11) Rabbit mAb #9733 using the CUT&Tag Assay Kit #77552. DNA libraries were prepared using the CUT&Tag Dual Index Primers and PCR Master Mix for Illumina Systems #47415. The figures show binding across the HOXA gene cluster (upper) and HOXD gene cluster (lower), known target genes of H3K27me3.
The CUT&Tag assay was performed with NCCIT cells and Ezh2 (D2C9) XP® Rabbit mAb #5246 using the CUT&Tag Assay Kit #77552. DNA libraries were prepared using the CUT&Tag Dual Index Primers and PCR Master Mix for Illumina Systems #47415. The figures show binding across the HOXA gene cluster (upper), and the HOXD gene cluster (lower), known target genes of Ezh2.
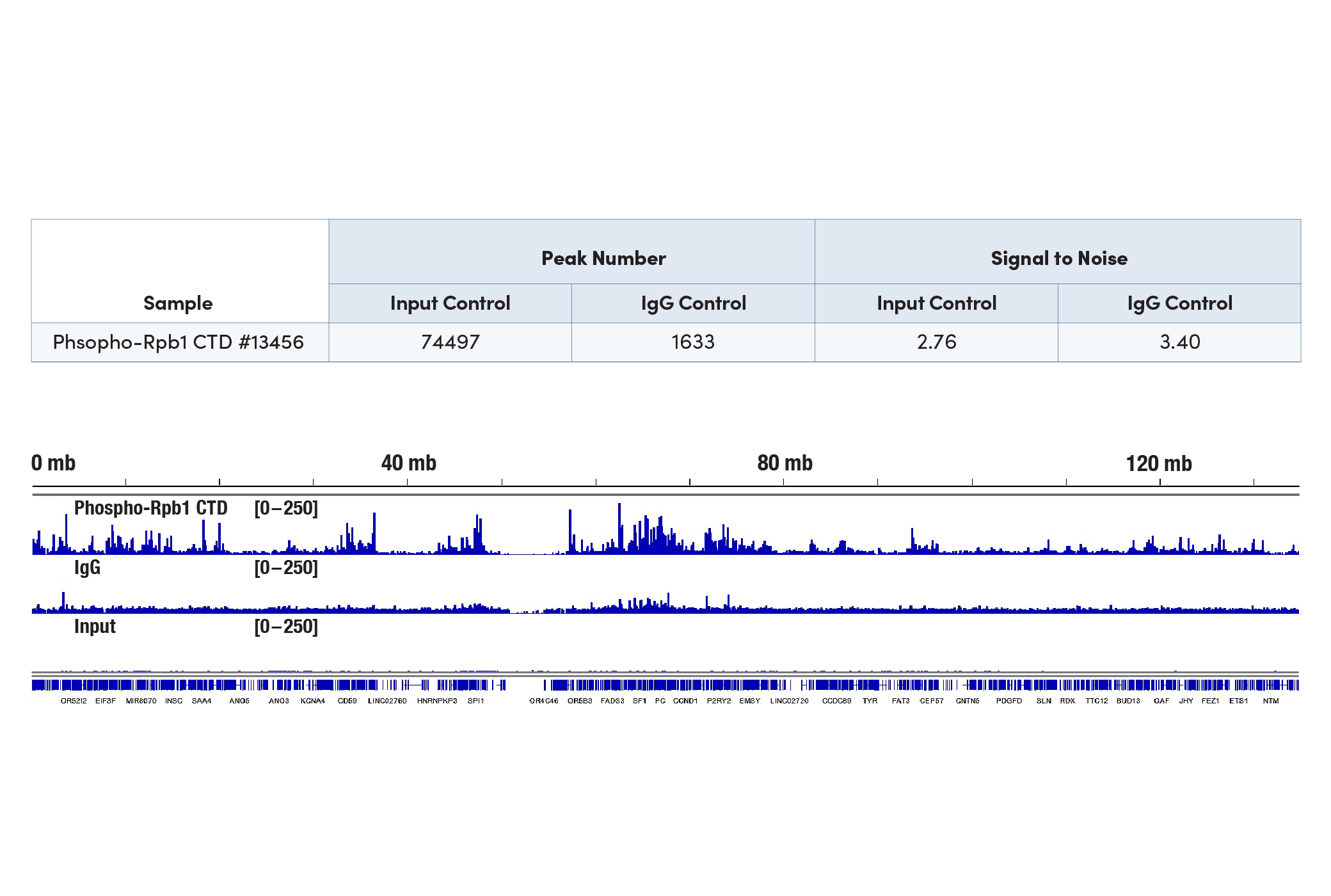
What controls can I use to show my CUT&Tag experiment is working?
We always recommend using a CUT&Tag-validated positive control antibody, like our Tri-Methyl-Histone H3 (Lys4) (C42D8) Rabbit mAb #9751, in your experiment to confirm that your CUT&Tag assay is working regardless of the performance of your desired target-specific antibody. For the negative control, either the IgG antibody or the input sample can be used in the CUT&Tag assay, depending on the requirements of the peak calling algorithm to be used. Both negative controls have their own advantages and limitations. The input DNA sample is fragmented genomic DNA that is not tagmented by Tn5, so it requires additional bench work compared to the CUT&Tag samples from the same experiment, including sonication fragmentation and in-vitro adaptor ligation steps for library preparation. The input DNA sample will always generate enough library yield and sequencing reads for data analysis. CUT&Tag signals are so strong, however, that some background signal can end up being counted as peaks if you are using the input DNA sample as the negative control. This issue can be avoided by using an IgG control because it performs better with regard to reasonable and reliable peak calling results (see images). However, using an IgG control runs the risk of obtaining a lower library yield with significantly fewer sequencing reads for your data analysis. Note that some labs analyze CUT&Tag peaks without any negative controls. All of the Tri-Methyl-Histone H3 (Lys4) (C42D8) Rabbit mAb #9751, Normal Rabbit IgG #2729, and Normal Mouse IgG #68860 control antibodies are included in our CUT&Tag Assay Kit #77552 for your convenience.
The CUT&Tag assay was performed with 100,000 HeLa cells and either Phospho-Rpb1 CTD (Ser2/Ser5) (D1G3K) Rabbit mAb #13546 or Normal Rabbit IgG #2729, using the CUT&Tag Assay Kit #77552. The DNA input sample was prepared following section VI of the CUT&RUN
Assay Kit #86652 protocol. DNA libraries were prepared using the CUT&Tag Dual Index Primers and PCR Master Mix for Illumina Systems #47415 for Rpb1 and IgG samples, and the DNA Library Prep Kit for Illumina Systems (ChIP-seq, CUT&RUN) #56795 for the input DNA sample. The figure shows enrichment across chromosome 11, indicating that the IgG sample identifies more background signal than the input sample and generates a more reasonable peak number accordingly (see table).
What is the best way to choose an antibody for my CUT&Tag assay if I can not find a CUT&Tag-validated antibody for my target protein?
We always recommend starting with CST CUT&Tag-validated antibodies because CST scientists perform intensive in-house tests to exclude any antibody that tends to show non-specific tagmentation at open chromatin regions (a unique problem for the CUT&Tag assay). If a CUT&Tag-validated antibody is not available, we recommend starting with a CUT&RUN-validated antibody. Alternatively, you may want to try an antibody validated for ChIP, ChIP-seq, or an immunofluorescence assay. The antibody binding in CUT&Tag takes place in an intact nuclear environment, resembling conditions for antibody binding in immunofluorescence assays. If a non-CUT&Tag-validated antibody is used, it is imperative to confirm that there is no non-specific tagmentation, otherwise your results might not be reliable.
Which method is better to use for purifying CUT&Tag DNA, a phenol-chloroform DNA extraction using ethanol precipitation or DNA affinity columns?
We recommend using CST DNA Purification Buffers and Spin Columns (ChIP, CUT&RUN, CUT&Tag) #14209 when purifying your CUT&Tag DNA (also included in CUT&Tag Assay Kit #77552). Although a method using phenol-chloroform followed by ethanol precipitation recovers DNA fragments of all sizes, CST DNA spin columns effectively recover DNA fragments greater than, or equal to, 35 bp, while the size of tagmented genomic DNA fragments is no less than 70 bp. Therefore, CST DNA spin columns provide a simple, inexpensive, and robust method for purifying your CUT&Tag DNA fragments.
Can CUT&Tag-enriched DNA be analyzed by qPCR, or do I have to do NGS to analyze my data?
If qPCR analysis is desired, we recommend performing CUT&RUN. CUT&Tag DNA is not compatible with qPCR. The sample incubation step at 58°C required for tagmented DNA solubilization in the CUT&Tag protocol breaks open the nuclear membrane, resulting in the final CUT&Tag DNA sample being composed of both target-bound tagmented DNA and background genomic DNA. If qPCR is performed with primers against target genes on the final CUT&Tag DNA sample, there is no way to distinguish the CUT&Tag signal from the background signal. However, qPCR analysis of target genes is possible using a CUT&Tag DNA library, because during the library amplification process the tagmented DNA fragments are selectively enriched while the background genomic DNA is diluted out. The qPCR analysis on a CUT&Tag DNA library can serve as a QC step before NGS. We recommend using the CUT&Tag Dual Index Primers and PCR Master Mix for Illumina Systems #47415 for CUT&Tag DNA library preparation.
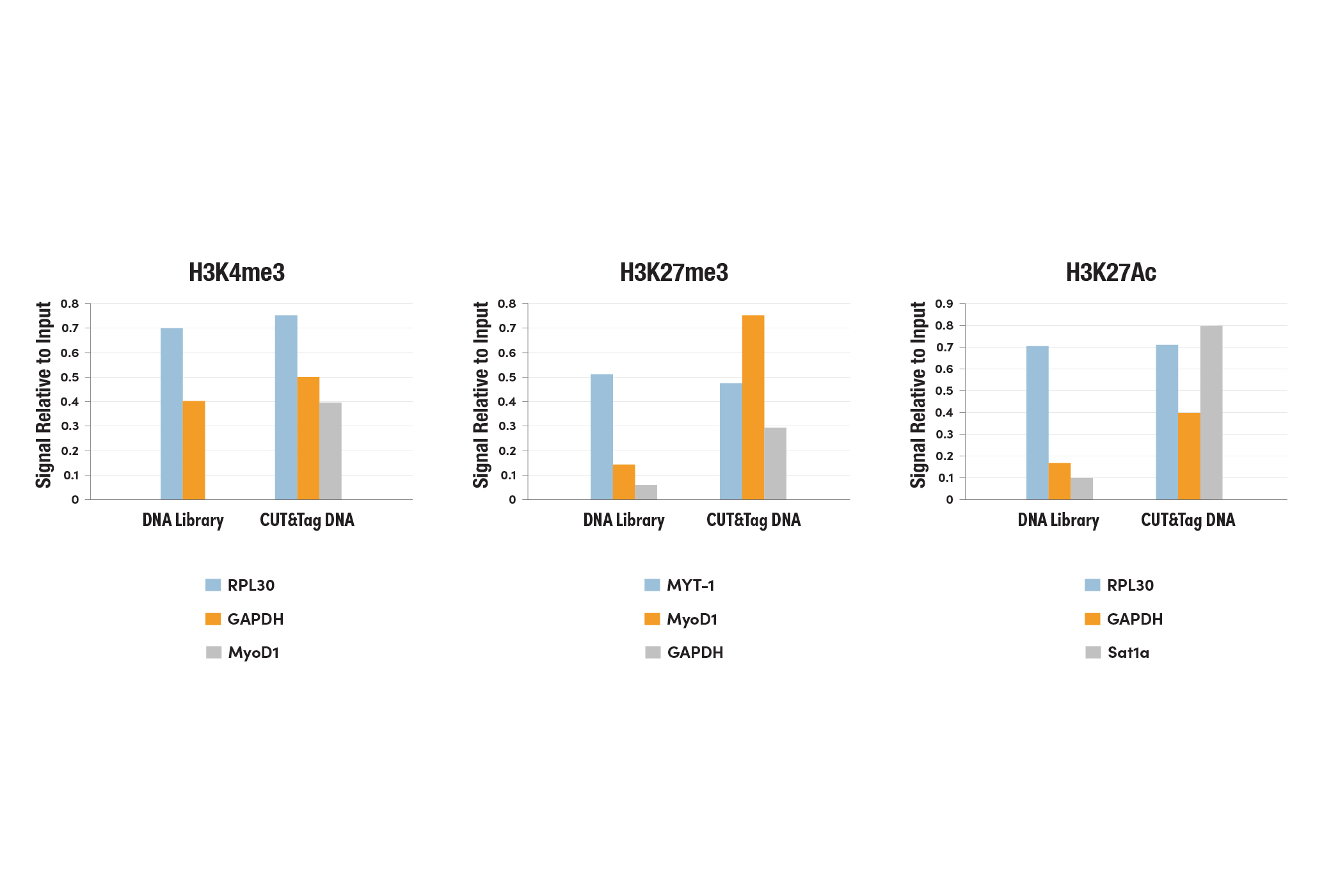
Left Panel: The CUT&Tag assay was performed with HeLa cells and Tri-Methyl-Histone H3 (Lys4) (C42D8) Rabbit mAb #9751, using the CUT&Tag Assay Kit #77552. DNA libraries were prepared using the CUT&Tag Dual Index Primers and PCR Master Mix for Illumina Systems #47415. The enriched DNA was quantified by real-time PCR using positive primer sets SimpleChIP® Human RPL30 Exon 3 Primers #7014 and human GAPDH exon 1 primers, and the negative primer set SimpleChIP® Human MyoD1 Exon 1 Primers #4490. The amount of immunoprecipitated DNA in each sample is represented as signal relative to the total amount of input chromatin, which is equivalent to one. Middle Panel: The CUT&Tag assay was performed with HeLa cells and Tri-Methyl-Histone H3 (Lys27) (C36B11) Rabbit mAb #9733, using the CUT&Tag Assay Kit #77552. DNA libraries were prepared using the CUT&Tag Dual Index Primers and PCR Master Mix for Illumina Systems #47415. The enriched DNA was quantified by real-time PCR using positive primer sets SimpleChIP® Human MyoD1 Exon 1 Primers #4490 and human MYT-1 exon 1 primers, and the negative primer set human GAPDH exon 1 primers. The amount of immunoprecipitated DNA in each sample is represented as signal relative to the total amount of input chromatin, which is equivalent to one.
Right Panel: The CUT&Tag assay was performed with HeLa cells and Acetyl-Histone H3 (Lys27) (D5E4) XP® Rabbit mAb #8173, using the CUT&Tag Assay Kit #77552. DNA libraries were prepared using the CUT&Tag Dual Index Primers and PCR Master Mix for Illumina Systems #47415. The enriched DNA was quantified by real-time PCR using positive primer sets SimpleChIP® Human RPL30 Exon 3 Primers #7014 and human GAPDH exon 1 primers, and the negative primer set SimpleChIP® Human α Satellite Repeat Primers #4486. The amount of immunoprecipitated DNA in each sample is represented as signal relative to the total amount of input chromatin, which is equivalent to one.
What is the expected yield of CUT&Tag DNA libraries?
The expected yield of an amplified CUT&Tag DNA library depends on the DNA quantification system used. If using a Nanodrop or QIAxpert System, the typical reading is 10-20 ng/µl for histone targets and 5-12 ng/µl for non-histone targets. If the library concentration is lower than 3 ng/µl with the Nanodrop or QIAxpert System, please refer to the troubleshooting guide before sequencing your samples. If using Qubit Fluorometric Quantification system or Picogreen assay, the typical reading is 3-10 ng/µl for histone targets and could be lower than 1 ng/µl for non-histone targets. Low yield doesn't necessarily indicate a failure of the CUT&Tag experiment. We would suggest continuing with NGS if the positive control Tri-Methyl-Histone H3 (Lys4) (C42D8) Rabbit mAb #9751 generates the expected library yield or the qPCR QC of the DNA library generates a good signal-to-noise ratio. Don't let the low yield stop you from obtaining quality NGS data.
I would like to QC and analyze the fragment sizes of my CUT&Tag DNA before sequencing, but my yield is too low to see on an Agilent Bioanalyzer or TapeStation system. What should I do?
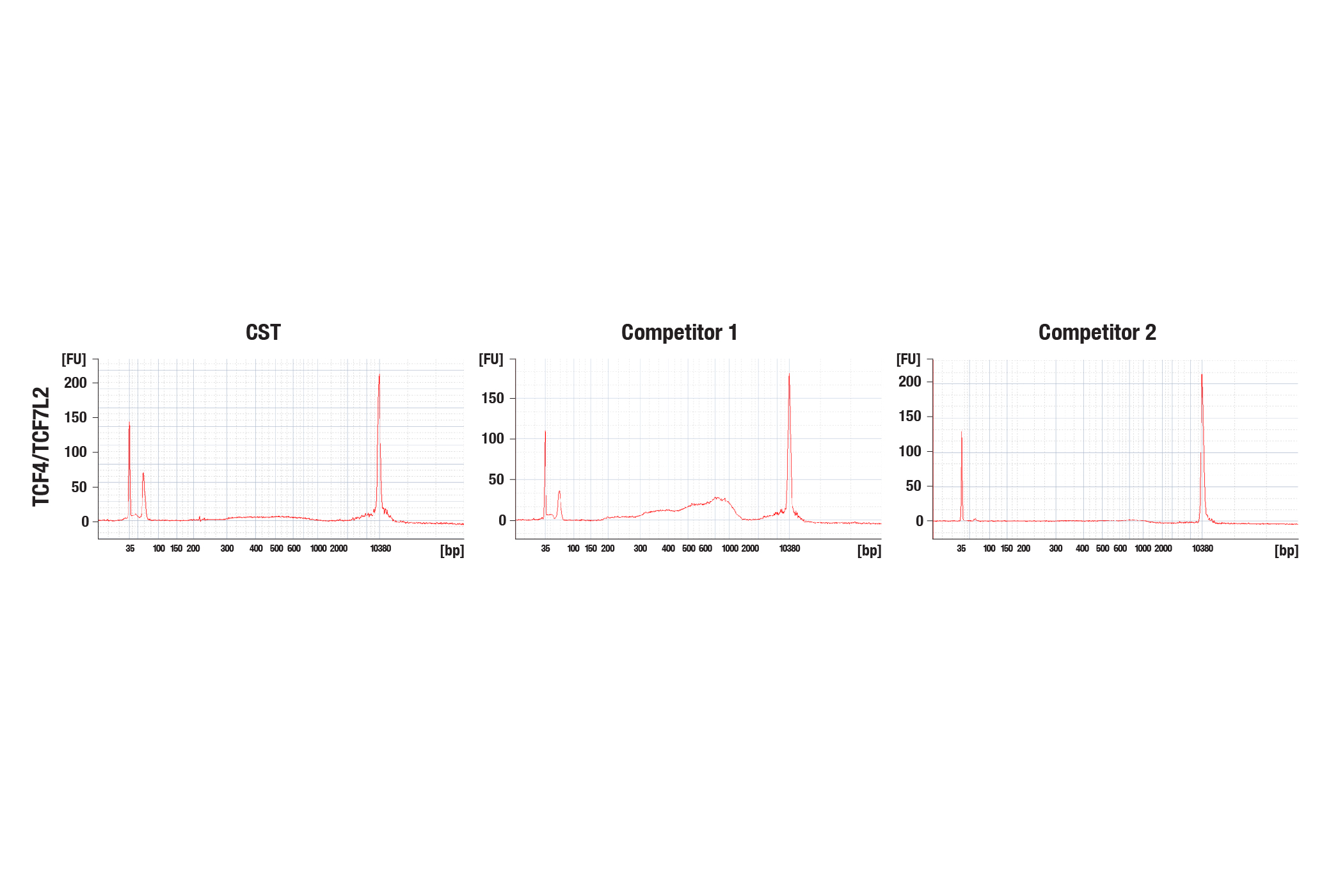
Due to the expected and typically low yield of CUT&Tag DNA, we recommend using all 30 µL of the CUT&Tag DNA sample (from section VI of the CUT&Tag protocol #77552) for library amplification. While CUT&Tag DNA libraries generated for histone modifications typically show some signal on the Agilent Bioanalyzer or TapeStation systems, libraries generated for non-histone proteins such as transcription factors and cofactors often have very weak or even no visible signal on Bioanalyzer systems, but they still generate NGS results with high-mapping rates, high numbers of identified binding peaks, and decent signal-to-noise ratios across the whole genome (see images). We find that the High Sensitivity ScreenTape Assay, rather than the Standard ScreenTape Assay, on the TapeStation system provides a better method for CUT&Tag library quality assessment due to its optimized fluorescent chemistry and reduced baseline noise. For CUT&Tag libraries where the Bioanalyzer or the TapeStation systems cannot accurately determine the average library size, we recommend using an estimated size of 900 bp. This allows low-yield libraries to be intentionally pooled at a slightly higher proportion than normal yield libraries.
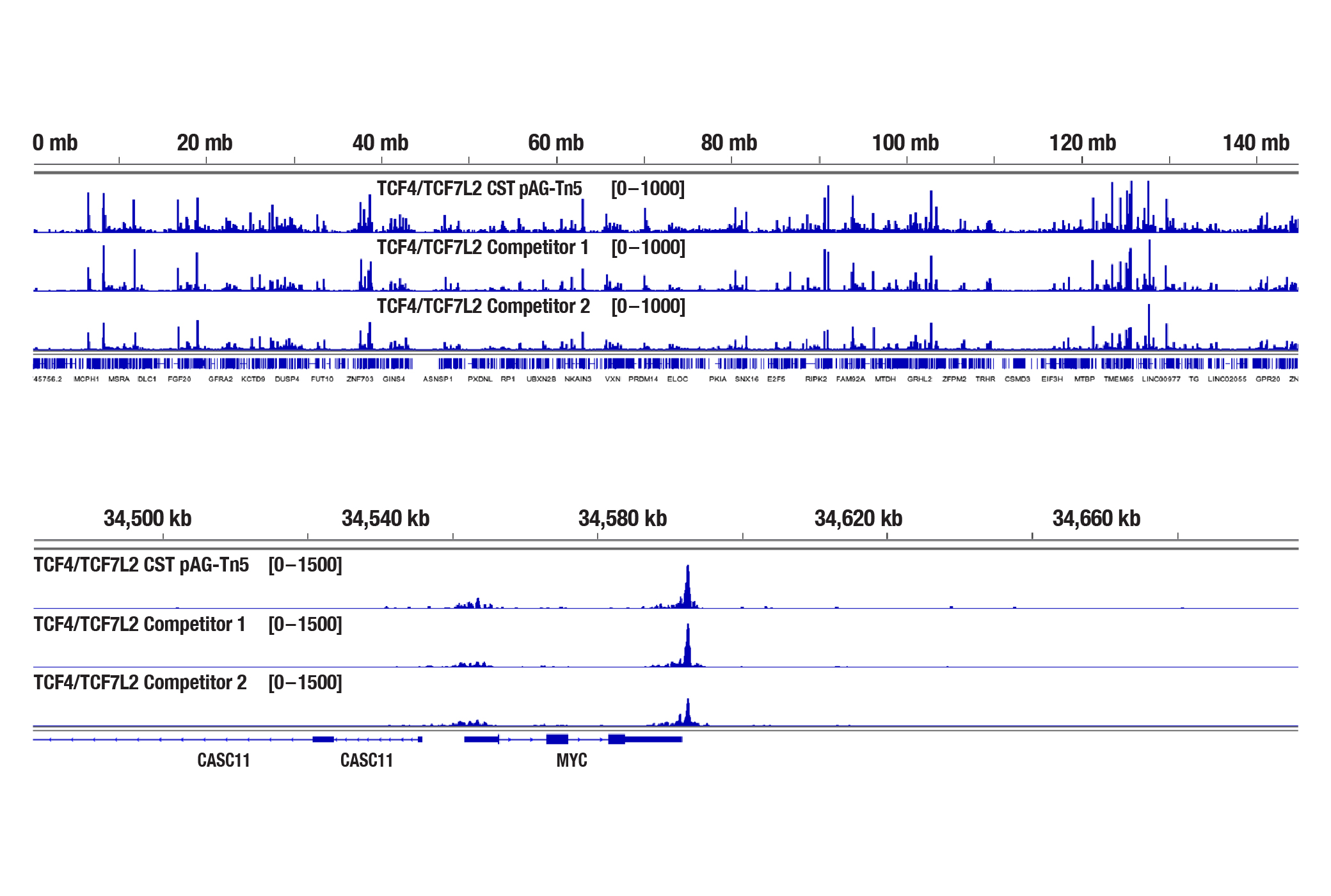
The CUT&Tag assay was performed with HCT 116 cells and TCF4/TCF7L2 (C48H11) Rabbit mAb #2569, with several sources of loaded pAG-Tn5. The amount of each enzyme used was based on the manufacturer's recommendation. DNA libraries were prepared using the CUT&Tag Dual Index Primers and PCR Master Mix for Illumina #47415. The figure (upper image) shows the profile obtained from a Bioanalyzer system for the CUT&Tag library DNA. The same DNA libraries analyzed with the Bioanalyzer system were sequenced and NGS tracks are shown (lower image). NGS data shows equivalent binding across chromosome 8 (upper), including MYC (lower), a known target gene of TCF4/TCF7L2.
The same DNA libraries analyzed with the Bioanalyzer system were sequenced and NGS tracks are shown (lower image). NGS data shows equivalent binding across chromosome 8 (upper), including MYC (lower), a known target gene of TCF4/TCF7L2.
How do I pool CUT&Tag DNA libraries with a variety of yields?
Usually the CUT&Tag DNA libraries from histone targets have a higher concentration than those from non-histone targets. We use the following formula to convert a library concentration from ng/µL to nM before diluting each library sample to the same concentration (nM) for pooling purposes: Concentration (nM) = 1,000,000 X Concentration (ng/µL) / library average size (bp) / 660. In addition to obtaining a greater library size for undetectable library samples from the Bioanalyzer or TapeStation system (as described in the above question), we would also suggest pooling the libraries that have a flat signal of 5-10 fold more than the libraries that show normal sized peaks on the Bioanalyzer or TapeStation systems. This ensures an even distribution of the number of reads among all samples. Usually, a library pool concentration of 2 nM DNA is enough for NGS purposes, although a higher concentration is always welcome.
What DNA library prep kit do you recommend using for the CUT&Tag assay?
We recommend using our CUT&Tag Dual Index Primers and PCR Master Mix for Illumina Systems #47415 to make DNA libraries for CUT&Tag. Our DNA Library Prep Kit for Illumina (ChIP-seq, CUT&RUN) #56795, and its paired Multiplex Oligos for Illumina (Dual Index Primers) (ChIP-seq, CUT&RUN) #47538 or Multiplex Oligos for Illumina (Single Index Primers) (ChIP-seq, CUT&RUN) #29580 are not compatible with CUT&Tag DNA samples.
Do you recommend performing size selection of the DNA during the library preparation?
No, we don't recommend performing size selection during the library purification for CUT&Tag. Based on our experience and feedback from customers, size selection can significantly decrease both the yield and diversity of the DNA library. This is extremely detrimental for CUT&Tag libraries due to the typical yield of the CUT&Tag assay. If you are concerned that large background DNA fragments (> 1 kb) will negatively impact the NGS, then simply decrease the extension time when amplifying the tagmented DNA during the library preparation. We find that reducing the extension time to 10-15 seconds greatly reduces the amplification of large fragments > 1kb, while still being sufficient for the amplification of smaller and more desirable DNA fragments during the CUT&Tag library preparation.
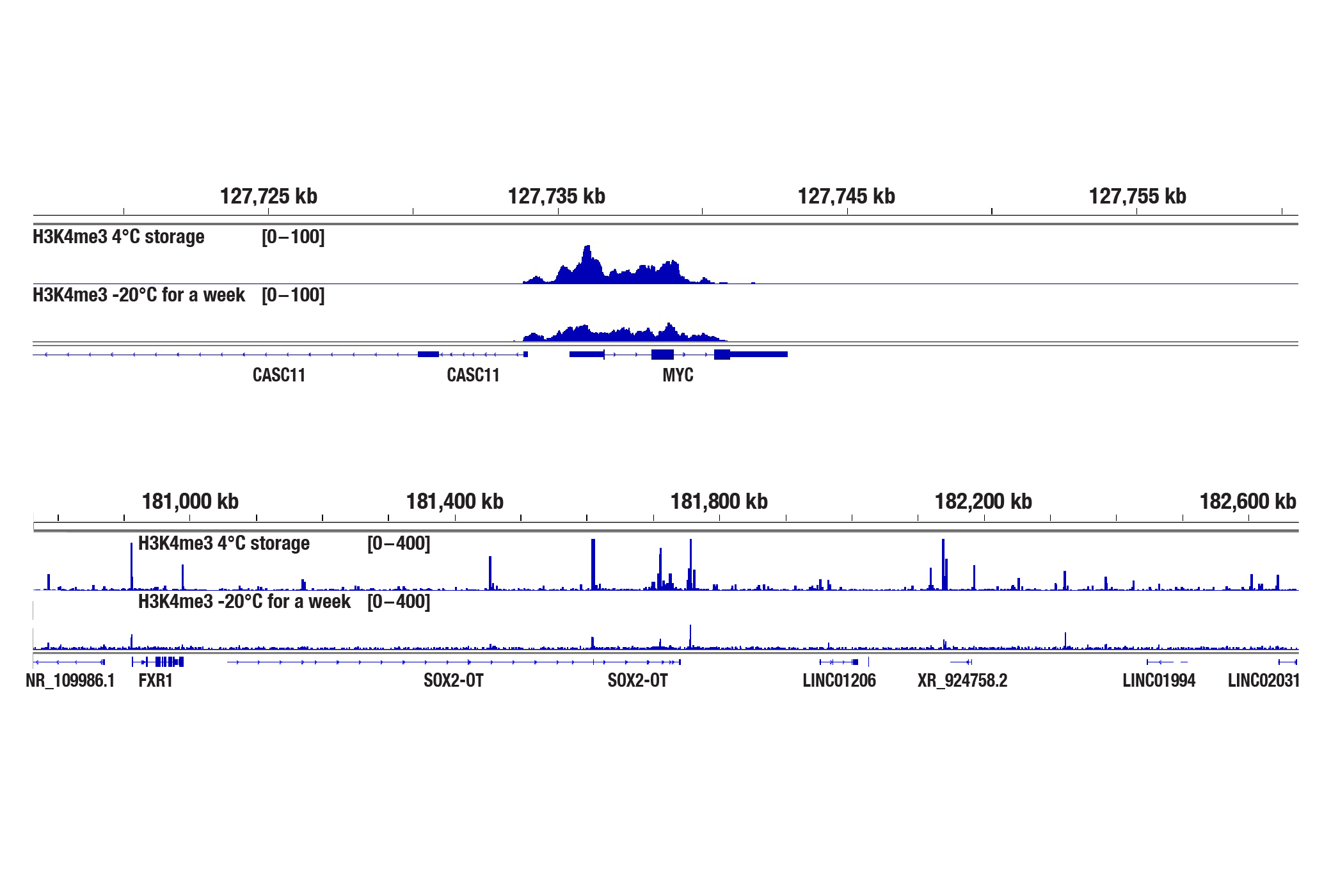
What if the whole CUT&Tag kit is left at -20°C or room temperature upon receipt?
The CUT&Tag Assay Kit #77552 contains components that are packaged carefully and need to be stored at either RT, 4°C, or -20°C. It's important to store each reagent at the proper temperature to ensure proper functionality. If the whole kit is accidentally left at -20°C, the Concanavalin A beads will lose some activity (see images). If this happens we recommend purchasing our standalone Concanavalin A Magnetic Beads and Activation Buffer #93569 for future experiments, especially when working with low abundance and/or weak DNA binding proteins. In addition, please thaw the DNA spin columns slowly (eg. thaw at 4°C first then calibrate to room temperature) and make sure there is no condensation inside the column before use. If needed, DNA Purification Buffers and Spin Columns (ChIP, CUT&RUN, CUT&Tag) #14209 are also available as a standalone product. All of the other reagents in the kit are fine to use after thawing. If the whole kit is left at room temperature for less than a week, the majority of the reagents will still be good to use, including the pAG-Tn5 (Loaded) and the Concanavalin A beads. Exceptions are the Spermidine solution and Protease Inhibitor Cocktail, which may have impaired performances. Both the 100X Spermidine #27287 and Protease Inhibitor Cocktail (200X) #7012 are available as standalone products.
The CUT&Tag assay was performed with 100,000 NCCIT cells, Concanavalin A beads stored at 4°C or frozen at -20°C for a week (as indicated), and either Tri-Methyl-Histone H3 (Lys4) (C42D8) Rabbit mAb #9751 (upper panel) or SMARCC1/BAF155 (D7F8S) Rabbit mAb #11956 (lower panel), using the CUT&Tag Assay Kit #77552. DNA libraries were prepared using the CUT&Tag Dual Index Primers and PCR Master Mix for Illumina Systems #47415. The upper panel shows H3K4me3 binding across MYC, a known target gene of H3K4me3. The lower panel shows SMARCC1 binding across Sox2, a known target gene of the BAF complex.
CST, Cell Signaling Technology, and SimpleChIP are registered trademarks of Cell Signaling Technology, Inc. All other trademarks are the property of their respective owners. Visit cellsignal.com/trademarks for more information.
CUT&Tag provided under a license from Active Motif, Inc. under U.S. Patent No. 10,689,643 and 9,938,524, foreign equivalents, and child patents deriving therefrom. For purchaser's internal research use only. May not be used for resale, services, or other commercial use.
U.S. Patent No. 11,733,248, foreign equivalents, and child patents deriving therefrom.
U.S. Patent No. 7,429,487, foreign equivalents, and child patents deriving therefrom.