Immuno-Parallel Reaction Monitoring (iPRM)
iPRM is a multiplexed LC/MS assay service that combines the specificity of mass spectrometry, the throughput of an immunoassay, and the power of multiplexing to accelerate the time-to-market of your drug and/or diagnostic test. Our iPRM services let you leverage the proteomic expertise of CST scientists and the established specificity and sensitivity of CST antibodies for streamlined assay development and reliable validation of your therapeutic target(s). iPRM services let you:
- Seamlessly transition from biomarker discovery to biomarker validation.
- Profile the impact of your drug across your choice of targets to determine on- and off-target effects.
- Quantify dozens of analytes in a single, complex biological sample with a high degree of confidence, sensitivity, and specificity.
Learn more about iPRM services or consult with a CST in-house expert.
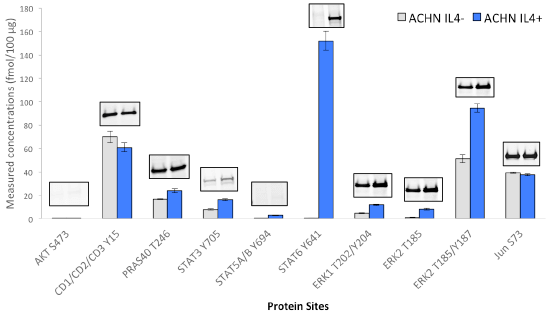
Immuno-PRM assay data and corresponding western blot results for phosphorylated targets in human renal carcinoma cell line ACHN under perturbation of Interleukin-4 (IL-4). The cells were treated with Human Interleukin-4 (hIL-4)#8919 at 40 ng/ml for 20 min before harvest. For iPRM assays, ACHN cell proteins were digested by trypsin, then 100 µg of ACHN tryptic peptides were mixed with heavy isotope labeled peptides, which served as internal standards. A multiplex immuno-precipitation was performed using 5 µg of each of the following CST antibodies: AKT S473 (#4060), CDK1/2/3 Y15 (#4539), PRAS40 T246 (#2997), STAT3 Y705(#9145), STAT5A/B Y694 (#9314), STAT6 Y641 (#9364), ERK1 T202/Y204(#4370), ERK2 T185(#4370), ERK2 T185/Y187 (#4370), Jun S73(#3270). Following immuno-enrichment, a multiplex PRM analysis of peptides was achieved in a Q Exactive™ mass spectrometer. Peak areas were then extracted in Skyline, and peptide concentrations were calculated based on peptide light-to-heavy ratios. Immuno-capture and PRM analysis were performed in triplicate. The same antibodies used for iPRM assays were also applied for western blotting. Twenty micrograms of protein extracts from control and treated ACHN cells were loaded on a 4% to 20% SDS-PAGE gradient gel for each blot. After transferred to nitrocellulose membrane, proteins were probed with phospho-specific antibodies at a dilution ratio of 1:1000.
How it works
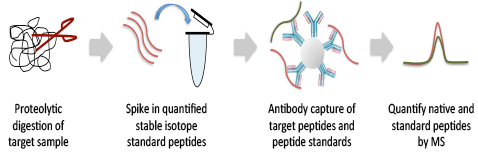
Lysates from a wide variety of research, clinical, or preclinical samples, including core needle biopsies, cells, or tissues, are proteolytically digested and assay targets are antibody-enriched along with quantified heavy-isotype peptide standards chemically identical to the targets of the assay. The enriched sample is analyzed on a Thermo Scientific™ Q Exactive™ Hybrid Quadrupole-Orbitrap™ Mass Spectrometer (MS) using a targeted PRM scan to precisely quantify the target and standard peptides with the highest sensitivity. Data are provided in Microsoft® Excel® and PowerPoint®.
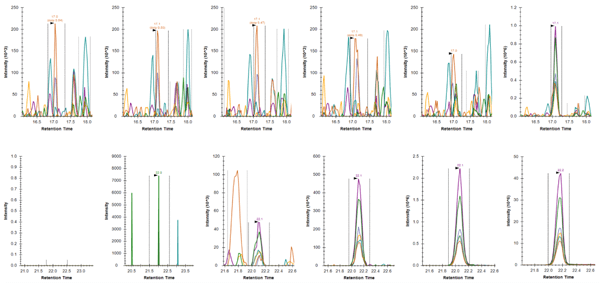
Importance of immuno-enrichment for PRM analysis
Comparison of Skyline-extracted peaks for various concentrations of phosphopeptide LAS*PELER++ from Jun (S73) after PRM (top row) and immuno-PRM (bottom row) analysis. The peaks of fragment ions (y7-98+, y6-98+, y5+, y3+, y7-98++, y6-98++) were extracted with mass tolerance 0.05 m/z from either direct injection of 1 µg trypsin digested SKBR3 lysate (top row; PRM) or from injection after immuno-precipitation of 100 µg SKBR3 peptides (bottom row; iPRM). The concentrations of the peptide in SKBR3 cell lysates were 0, 0.2, 2, 20, 200, 2000 amol/µg from left to right. The lower limit of detection (LLOD) for the peptide was enhanced from 2000 amol/µg using PRM method to 2 amol/µg using iPRM workflow.
Service options
Pick from our list of ready-to-go assays below. These assays are pre validated and use CST components, including the high-affinity antibody and in-house synthesized AQUA peptide standards.
| Catalog # | Protein Site | Peptide | LLOD (fmol/100 μg) | LLOQ (fmol/100 μg) |
|---|---|---|---|---|
| 4060 | Akt S473 | RPHFPQFS*YSASGTA | 0.059 | 0.117 |
| 4539 | CDK1/CDK2/CDK3 Y15 | IGEGTY*GVVYK | 0.015 | 0.046 |
| 2997 | PRAS40 T246 | LNT*SDFQK | 0.004 | 0.012 |
| 9145 | STAT3 Y705 | YCRPESQEHPEADPGSAAPY*LK | 0.124 | 0.373 |
| 9314 | STAT5A/B Y694 | AVDGY*VKPQIK | 0.033 | 0.100 |
| 9364 | STAT6 Y641 | GY*VPATIK | 0.018 | 0.055 |
| 4370 | ERK1 T202/Y204 | IADPEHDHTGFLT*EY*VATR | 0.047 | 0.142 |
| 4370 | ERK2 T185 | VADPDHDHTGFLT*EYVATR | 0.254 | 0.763 |
| 4370 | ERK2 T185/Y187 | VADPDHDHTGFLT*EY*VATR | 0.033 | 0.098 |
| 3270 | Jun S73 | LAS*PELER | 0.021 | 0.063 |
LLOD and LLOQ were determined based on limit of blank, variance of the blank sample and variance of the lowest level spike-in sample.
If we don’t have the assay you’re looking for yet, we’re happy to develop it. A CST proteomics expert will work with you to define a custom assay. CST then develops and validates the assay in-house. If you’re interested in a target we don’t currently offer, we can collaborate with you to develop a custom antibody with the specificity and sensitivity CST antibodies are known for. Externally developed antibodies can also be used.
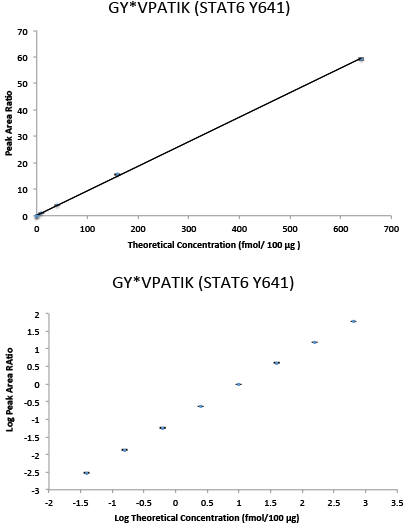
Assay Response Curve
Immuno-PRM response curve data in linear (top) and log space (bottom) for phospho-STAT6 (Y641) peptide GY*VPATIK. Multipoint experiments were performed by immunoprecipitating increasing amounts of 10 heavy isotope-labeled synthetic peptides in a background of 100 µg of tryptic peptides derived from the breast cancer cell line SKBR-3 with light peptides spiked in as internal standards. Following immunoprecipitation, peptides were analyzed on a Q Exactive™ Mass Sepctrometer. Peptide abundance was determined by extracting fragment peptide area in Skyline. Quantitation was achieved by calculating the ratio of peak areas (heavy/light) of the 3 to 6 abundant fragment ions for each peptide. The current figures show representative data for one phosphopeptide, while a 10-target multiplex PRM assay was performed. The error bars indicate the minimum and maximum of the peak area ratios of the three capture and PRM replicates. The response curve shows a linear range of over 4 orders of magnitude and an LLOQ of 0.039 fmol/100 µg for phosphopeptide GY*VPATIK.
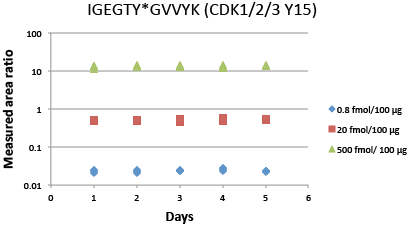
Assay Reproducibility
Immuno-PRM 5-day repeatability data for phosphopeptide IGEGTY*GVVYK from CDK1/2/3 (Y15). Samples are prepared by spiking 10 heavy isotope-labeled peptides at three levels (500 fmol, 20 fmol, 0.8 fmol as High, Med, Low level) in a background of 100 µg of tryptic peptides derived from the breast cancer cell line SKBR-3 with light peptides spiked in as internal standards. Triplicate enrichments were performed independently on 5 different days to measure repeatability. Following immunoprecipitation, peptides were analyzed on a Q Exactive™ Mass Spectrometer. Peak areas of 3 to 6 abundant fragments for each peptide were extracted in Skyline, and the peak area ratios were calculated based on the sum of fragments from light and heavy peptides. Average intra-assay and inter-assay CVs were measured for different starting concentrations of each peptide. The total CV was the square root of the sum of squares of intra- and inter-assay CVs. For phosphopeptide IGEGTY*GVVYK shown in the figure, all CVs are within 10%. For other peptides in the 10-target multiplex iPRM assay, all CVs are within 20%.
| Average intra-assay (within day) CV | Average inter-assay (between day) CV | Total CV √(intraCV²+interCV²) | n= | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| 6.0% | 5.2% | 7.5% | 7% | 7% | 5% | 9% | 9% | 9% | 15 | 15 | 15 |