PTMScan® Direct Cell Cycle & DNA Damage
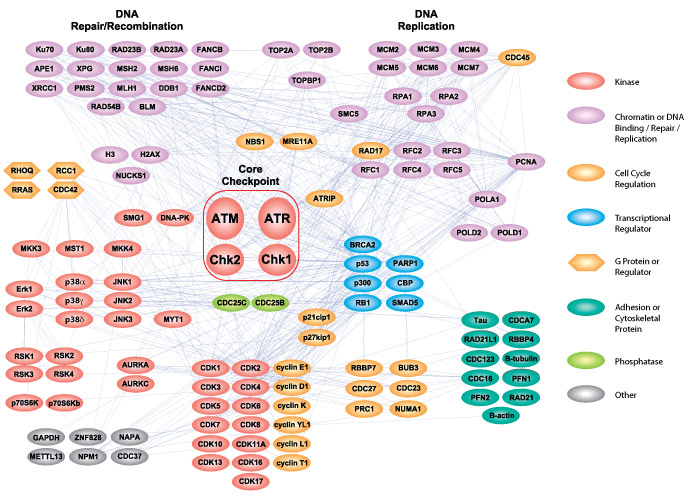
PTMScan® Direct Cell Cycle and DNA Damage Service allows access to perform targeted screening and quantification of a defined set of protein sites within the relevant DNA damage and cell cycle control pathways that are critical for drug discovery, biomarker discovery, or diagnostic development. The table below illustrates the list of protein sites that can be monitored with PTMScan® Direct Cell Cycle and DNA Damage Service.
Altogether, the PTMScan® Direct Cell Cycle & DNA Damage service provides a robust method to qualitatively identify and quantitatively profile 168 unique phosphorylation sites to 263 proteins.